For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NLQLFLLLIVVVTALAFDFTNGFHDTGNAMATSIASGALAPRVAVALPAVLNLIGAFLSTAVAATIAKGL IDANLVTLELVFAGLVGGIVWNLLTWLLGIPSSSSHALIGGIVGATIAAVGLRGVIWSGVVSKVIVPAVV AALLATLVGAVGTWLVYRTTRGVAEKRTERGFRRGQIGSASLVSLAHGTNDAQKTMGVIFLALMSYGAVS TTASVPPLWVIVSCAVAMAAGTYLGGWRIIRTLGKGLVEIKPPQGMAAESSSAAVILLSAHFGYALSTTQ VATGSVLGSGVGKPGAEVRWGVAGRMVVAWLVTLPLAGLVGAFTYGLVHFIGGYPGAILGFALLWLTATA IWLRSRRAPIDHTNVNADWEGNLTAGLEAGAQPLADQRPPVPAPPAPTPPPNHRAPQFGVTTRNAP*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
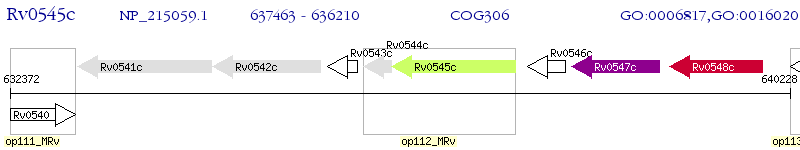
| M. tuberculosis H37Rv | Rv0545c | pitA | - | 100% (417) | PROBABLE LOW-AFFINITY INORGANIC PHOSPHATE TRANSPORTER INTEGRAL MEMBRANE PROTEIN PITA |
| M. tuberculosis H37Rv | Rv2281 | pitB | 6e-20 | 32.97% (185) | phosphate ABC transporter permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0559c | pitA | 0.0 | 99.76% (417) | inorganic phosphate transporter |
| M. gilvum PYR-GCK | Mflv_0031 | - | 1e-129 | 57.31% (417) | phosphate transporter |
| M. leprae Br4923 | MLBr_02260 | pitA | 1e-179 | 76.56% (418) | low-affinity inorganic phosphate transporter |
| M. abscessus ATCC 19977 | MAB_3950c | - | 1e-132 | 55.32% (432) | inorganic phosphate transporter |
| M. marinum M | MMAR_0891 | pitA | 0.0 | 84.78% (414) | low-affinity inorganic phosphate transporter, PitA |
| M. avium 104 | MAV_4599 | - | 0.0 | 80.00% (405) | phosphate transporter |
| M. smegmatis MC2 155 | MSMEG_1064 | - | 1e-129 | 56.97% (416) | phosphate/sulphate permease |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2694 | pitB | 5e-17 | 30.77% (182) | phosphate-transport permease PitB |
| M. vanbaalenii PYR-1 | Mvan_0939 | - | 1e-131 | 56.94% (425) | phosphate transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0545c|M.tuberculosis_H37Rv -------------------------------------------------M
Mb0559c|M.bovis_AF2122/97 -------------------------------------------------M
MMAR_0891|M.marinum_M -------------------------------------------------M
MLBr_02260|M.leprae_Br4923 -------------------------------------------------M
MAV_4599|M.avium_104 -------------------------------------------------M
Mflv_0031|M.gilvum_PYR-GCK ------------------------MGSISHPPTPAKIRKTGAEGRRINSM
MSMEG_1064|M.smegmatis_MC2_155 -------------------------------------------------M
Mvan_0939|M.vanbaalenii_PYR-1 -------------------------------------------------M
MAB_3950c|M.abscessus_ATCC_199 -------------------------------------------------M
MUL_2694|M.ulcerans_Agy99 MASGLQDPITPTPQYGGLFGQDRAWHLSFGLLLAGSFVLFSFWAFDYAGP
Rv0545c|M.tuberculosis_H37Rv NLQLFLLLIVVVTALAFDFTNGFHDTGNAMATSIASGALAPRVAVALPAV
Mb0559c|M.bovis_AF2122/97 NLQLFLLLIVVVTALAFDFTNGFHDTGNAMATSIASGALAPRVAVALSAV
MMAR_0891|M.marinum_M NIDLFLLIIVVITALAFDFTNGFHDTGNAMATSIASGALAPRTAVILSAA
MLBr_02260|M.leprae_Br4923 NINLFLLIIVVITALAFDFTNGFHDTGNAMATSIASGALAPKVAVFFSAI
MAV_4599|M.avium_104 NIQLFLLIIVVITALAFDFTNGFHDTGNAMATSIASGALKPKTAVALSAV
Mflv_0031|M.gilvum_PYR-GCK SAELVVLVLLIITALAFDFTNGFHDTGNAMATSIATGALKPKTAVLLAGI
MSMEG_1064|M.smegmatis_MC2_155 SAELVVLVLLIATALAFDFTNGFHDTGNAMATSIATRALKPKTAVLLAGV
Mvan_0939|M.vanbaalenii_PYR-1 SAELIILVLLIATALAFDFTNGFHDTGNAMATSIATGALKPKTAVILAGV
MAB_3950c|M.abscessus_ATCC_199 DATMIILVLLIATALAFDFTNGFHDTGNAMATSIATGALKPKTAVLLAGI
MUL_2694|M.ulcerans_Agy99 GANKLILVLATVVGMFMAFNVGGNDVANSFGTSVGAGTLSMKQALLVAAI
. .:*:: ..: : *. * :*..*::.**:.: :* : *: ...
Rv0545c|M.tuberculosis_H37Rv LNLIGAFLS-TAVAATIAKGLIDAN-------------LVTLELVFAGLV
Mb0559c|M.bovis_AF2122/97 LNLIGAFLS-TAVAATIAKGLIDAN-------------LVTLELVFAGLV
MMAR_0891|M.marinum_M LNLLGAFLS-TAVAATIAKGLIDAN-------------LVTLELVFAGLV
MLBr_02260|M.leprae_Br4923 LNLVGAFLS-TAVAATIAKDLIEAD-------------LVTLELVFAGLV
MAV_4599|M.avium_104 LNLVGAFMS-TAVAATIAKGLIDSN-------------IVTLELVFAGLV
Mflv_0031|M.gilvum_PYR-GCK LNLVGAFLS-VEVAVTVTTSVLKVQDSTSGELLSGIDASTGLTIIFAGLI
MSMEG_1064|M.smegmatis_MC2_155 LNLVGAFLS-VEVAITVTSSVLKIQDSSTGQLVGSIDASTGLTIIFAGLI
Mvan_0939|M.vanbaalenii_PYR-1 LNLVGAFLS-VEVAVTVTTSVLKIQDSKTGQLIPSIDASTGLTIIFAGLI
MAB_3950c|M.abscessus_ATCC_199 LNLVGAFLS-VEVAVTVTTSVIKVQDSKTGHLLPNITPSMGLTIIFAGLI
MUL_2694|M.ulcerans_Agy99 FEVSGAVIAGGDVTETIRSGIVDLSG-------VSVAPNDFMNIMLAALL
::: **.:: *: *: ..::. . : :::*.*:
Rv0545c|M.tuberculosis_H37Rv GGIVWNLLTWLLGIPSSSSHALIGGIVGATIAAVGLRG---VIWSGVVSK
Mb0559c|M.bovis_AF2122/97 GGIVWNLLTWLLGIPSSSSHALIGGIVGATIAAVGLRG---VIWSGVVSK
MMAR_0891|M.marinum_M GGIVWNLLTWLLGIPSSSSHALIGGIVGATIAAVGGRG---VIWSGVVSK
MLBr_02260|M.leprae_Br4923 GGIVWNLLTWLLGIPSSSSHALIGGIVGARIAAVGGHG---VIWSGVISK
MAV_4599|M.avium_104 GGVVWNLLTWLLGIPSSSSHALIGGIVGATIAAVGAHG---VIWKGVISK
Mflv_0031|M.gilvum_PYR-GCK GGILWNLLTWLFGIPSSSSHALFGGLIGAGLAALGLAG---VNWSGVTQK
MSMEG_1064|M.smegmatis_MC2_155 GGILWNLLTWLFGIPSSSSHALFGGLIGAGLAAIGLAG---VNWAGVTQK
Mvan_0939|M.vanbaalenii_PYR-1 GGILWNLLTWLFGIPSSSSHALFGGLIGAGLAAVGLAG---VNWSGVTQK
MAB_3950c|M.abscessus_ATCC_199 GGILWNLLTWLFGIPSSSSHALFGGLIGAALAAIGTSG---VKWDGILQK
MUL_2694|M.ulcerans_Agy99 AAAMWLLFANRMGYPVSTTHSIVGGIVGAAVTLGIVDGKGLSALQMVQWD
.. :* *:: :* * *::*::.**::** :: * : .
Rv0545c|M.tuberculosis_H37Rv VIVPAVVAALLATLVGAVGTWLVYRTTRGVAEKRTERGFRRGQIGSASLV
Mb0559c|M.bovis_AF2122/97 VIVPAVVAALLATLVGAVGTWLVYRTTRGVAEKRTERGFRRGQIGSASLV
MMAR_0891|M.marinum_M VIVPAVVAALLATLVGAIGTWLVYRITATVSEKDTERGFRRGQIGSASLV
MLBr_02260|M.leprae_Br4923 VIIPAIIAALLAIVVGAVATWLVYAITRSVPAMSTDTRFRRGQIGSASLV
MAV_4599|M.avium_104 AIIPAIVSAILAILVGAVATWLVYRITRGVPKKRTEAGFRRGQIGSASLV
Mflv_0031|M.gilvum_PYR-GCK VLIPALAAPMIACLVAGCGTWLVYRLTRRVAEKRRREGFRWGQIATASLV
MSMEG_1064|M.smegmatis_MC2_155 VLIPAVAAPVIACLVASCGTWLIYRITRNVAQKRREQGFRVGQIATASLV
Mvan_0939|M.vanbaalenii_PYR-1 VLIPAVAAPLIACLVAGCGTWLVYRITRNVMKNRREQGFRWGQIATASLV
MAB_3950c|M.abscessus_ATCC_199 VIIPAVAAPLIAGLVAAAGTWLVYRITRNVVKKRREEGFRWGQIATASLV
MUL_2694|M.ulcerans_Agy99 EIGQIAISWVLSPLLGGLVSYALYGAIKRHILLYNEEAERKLLEIKKERI
: : ::: ::.. :: :* * . . :
Rv0545c|M.tuberculosis_H37Rv SLAHGTNDAQKTMGVIFLALMSYGAVSTTASVP--PLWVIVSCAVAMAAG
Mb0559c|M.bovis_AF2122/97 SLAHGTNDAQKTMGVIFLALMSYGAVSTTASVP--PLWVIVSCAVAMAAG
MMAR_0891|M.marinum_M SLAHGTNDAQKTMGVIFLALMSYGAVSTSATMP--PLWVIVSCALAMAGG
MLBr_02260|M.leprae_Br4923 SLAHGTNDAQKTMGVIFLALMSYGTVSKTASTP--PLWVIVCCAIAIAAG
MAV_4599|M.avium_104 SLAHGTNDAQKTMGIIFLALISYGSVSKTAAMP--PLWVIVSCALAMATG
Mflv_0031|M.gilvum_PYR-GCK ALSHGTNDAQKTMGVIALALITTGHLTGDVKEEGLPFWIIFSCAVAIGLG
MSMEG_1064|M.smegmatis_MC2_155 ALSHGTNDAQKTMGVIALALITTGHLSGDVKENGLPFWIIASCALAIGLG
Mvan_0939|M.vanbaalenii_PYR-1 ALSHGTNDAQKTMGVIALALITTGHLTGDVKEQGLPIWIIASCALAIGLG
MAB_3950c|M.abscessus_ATCC_199 ALSHGTNDAQKTMGVIALALITTGHLTGNVKETGLPFWIIASCALAIGLG
MUL_2694|M.ulcerans_Agy99 AHRERHKAAFERLSEIQQIAYTGAFARDAVAANRRDFDPDELESDYYREL
: . : * : :. * : . . : :
Rv0545c|M.tuberculosis_H37Rv TYLGGWRIIRTLGKGLVEIKPPQGMAAESSSAAVILLSAHFGYALSTTQV
Mb0559c|M.bovis_AF2122/97 TYLGGWRIIRTLGKGLVEIKPPQGMAAESSSAAVILLSAHFGYALSTTQV
MMAR_0891|M.marinum_M TYLGGWRIIRTLGKGLVEIKPPQGMAAEASSAAVILLSAHFGYALSTTQV
MLBr_02260|M.leprae_Br4923 TYLGGWRIIRTLGKGMVEIKPPQGMAAESSSAAVILLSAHFGYALSTTQV
MAV_4599|M.avium_104 TYLGGWRIIRTLGKGLVEIQSPQGMAAESSSAAVILLSAHFGYALSTTQV
Mflv_0031|M.gilvum_PYR-GCK TYLGGWRVIRTLGKGLVEIQSPQGFAAEASSAAIILSSSAAGMALSTTHV
MSMEG_1064|M.smegmatis_MC2_155 TYLGGWRVIRTLGKGLVEIESPQGLAAEASSAAIILSSSAAGMALSTTHV
Mvan_0939|M.vanbaalenii_PYR-1 TYLGGWRVIRTLGKGLVEIESPQGLAAEASSAAIILSSSAAGMALSTTHV
MAB_3950c|M.abscessus_ATCC_199 TYLGGWRVIRTLGKGLVEIESPQGLAAEASSAAIILSSSAAGMALSTTHV
MUL_2694|M.ulcerans_Agy99 HEIDTKRSSVDAFRALQNWVPLVAAAGSMIISAMLLFKGFKHMHLGLTTM
:. * :.: : . . *.. :*::* .. *. * :
Rv0545c|M.tuberculosis_H37Rv ATGSVLGSG----------VGKPGAEVRWGVAGRMVVAWLVTLPLAG---
Mb0559c|M.bovis_AF2122/97 ATGSVLGSG----------VGKPGAEVRWGVAGRMVVAWLVTLPLAG---
MMAR_0891|M.marinum_M ATGSVLGSG----------VGKPGAEVRWGVATRMVAAWVITLPLAG---
MLBr_02260|M.leprae_Br4923 CTGSVLGSG----------LGKPGGEVRWGVAGRMATAWLVTLPLAG---
MAV_4599|M.avium_104 CTGSVLGSG----------LGKPGGEVRWGVAGRMGVAWLVTLPLAG---
Mflv_0031|M.gilvum_PYR-GCK ATGSILGSG----------VGKPGAEVRWAVAGRMAVAWLITLPAAG---
MSMEG_1064|M.smegmatis_MC2_155 ATGSILGSG----------VGKPGAEVRWAVAGRMAVAWLVTLPAAG---
Mvan_0939|M.vanbaalenii_PYR-1 ATGSILGSG----------VGKPGAEVRWAVAGRMAVAWLITLPAAG---
MAB_3950c|M.abscessus_ATCC_199 ATGSILGTG----------VGKPGAEVRWAVAGRMVLAWLVTLPAAG---
MUL_2694|M.ulcerans_Agy99 NNFFIMGMVGAAVWMATFILAKTLRGESLSRSTYLMFSWMQVFTASGFAF
. ::* :.*. . : : :*: .:. :*
Rv0545c|M.tuberculosis_H37Rv ---------LVGAFTYGLVHF-IGGYPGAILG-----FALLWLTATAIWL
Mb0559c|M.bovis_AF2122/97 ---------LVGAFTYGLVHF-IGGYPGAILG-----FALLWLTATAIWL
MMAR_0891|M.marinum_M ---------LVGAITYWLVHM-IGGYPGAIIG-----FALLCLVAATIWL
MLBr_02260|M.leprae_Br4923 ---------SVGAVTYWIVHL-IGGYPGAVIG-----FSLLVAASVAIYI
MAV_4599|M.avium_104 ---------LVGAVTYWIVHL-IGGYPGAIIG-----FALLVAVSATIYL
Mflv_0031|M.gilvum_PYR-GCK ---------LVGALTYWLSSG-VTSATGSDLAGDGLIFVILVGLSAFMWW
MSMEG_1064|M.smegmatis_MC2_155 ---------IVGALAFWLSHG-VERLTGSSLAGDGLIFMVLVGLSFYMWW
Mvan_0939|M.vanbaalenii_PYR-1 ---------IVGALSFWLSHG-AESLTGSSLAGDGLIFLLLCGLSFYLWW
MAB_3950c|M.abscessus_ATCC_199 ---------IVGALSYWLSHL-VGNLT-TPMVGDLIIFALLVVLSGYMWW
MUL_2694|M.ulcerans_Agy99 SHGSNDIANAIGPFAAVLDVLRSGSIEGKAAVPAAAMITFGVALCARLWF
:*..: : : . . ::
Rv0545c|M.tuberculosis_H37Rv RSR--RAPIDHTNVNADWEGNLTAGLEAGAQPLA----DQRPPVPAPPAP
Mb0559c|M.bovis_AF2122/97 RSR--RAPIDHTNVNADWEGNLTAGLEAGAQPLA----DQRPPVPAPPAP
MMAR_0891|M.marinum_M RSR--KARVDHTNVNAEWEGSLTAGLDGAPGPTAPSPSDRQRPPPPPPPP
MLBr_02260|M.leprae_Br4923 RSR--KVKVDHKNVNENWEGSLTAGLDGSDEHKP-------HSDVGPKLS
MAV_4599|M.avium_104 RSR--KVKVDHHNVNAEWKGDLTTGLEGADDHSP-------PPDAGPPFG
Mflv_0031|M.gilvum_PYR-GCK RAQ--QQKVDHKNVNADWDDTTNSVVPAELRVTT-------TPD------
MSMEG_1064|M.smegmatis_MC2_155 RAQ--QQKVDHNNVNADWDESTNSVVPADLKVTI-------PDNS-----
Mvan_0939|M.vanbaalenii_PYR-1 RAQ--QQKVDHSNVNADWDASTNSVVPAEVREAT-------TNGTPPTDT
MAB_3950c|M.abscessus_ATCC_199 RAQ--QEKVDSSNVNAEWDDSTNSVVPADVRESR-------AEAPVEAPN
MUL_2694|M.ulcerans_Agy99 IGRRVIATVGHNLTTMHPASGFAAELSAAGVVMGATVLGLPVSSTHILIG
.: :. .. . : : .
Rv0545c|M.tuberculosis_H37Rv TPPPNHRAPQFG----VTTRNAP-----------------------
Mb0559c|M.bovis_AF2122/97 TPPPNHRAPQFG----VTTRNAP-----------------------
MMAR_0891|M.marinum_M SPHSNGHTPRYGSDDALTAGNPS-----------------------
MLBr_02260|M.leprae_Br4923 ATLPRYRSSHHT----VGVRNAS-----------------------
MAV_4599|M.avium_104 GPGDRYQSDDPT----LKAS-AS-----------------------
Mflv_0031|M.gilvum_PYR-GCK -----PKPVATV----------------------------------
MSMEG_1064|M.smegmatis_MC2_155 ---ERPTPAGV-----------------------------------
Mvan_0939|M.vanbaalenii_PYR-1 KPPAGSKPAAAV----------------------------------
MAB_3950c|M.abscessus_ATCC_199 KQAPNKEAANKEAPKKDSANDTASV---------------------
MUL_2694|M.ulcerans_Agy99 AVLGVGIVNRATNWALMKPIALAWVIALPSAAILSAVGLVTLRAIF