For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDESSVRTPKALPDGVSQATVGVRGGMLRSGFEETAEAMYLTSGYVYGSAAVAEKSFAGELDHYVYSRY GNPTVSVFEERLRLIEGAPAAFATASGMAAVFTSLGALLGAGDRLVAARSLFGSCFVVCSEILPRWGVQT VFVDGDDLSQWERALSVPTQAVFFETPSNPMQSLVDIAAVTELAHAAGAKVVLDNVFATPLLQQGFPLGV DVVVYSGTKHIDGQGRVLGGAILGDREYIDGPVQKLMRHTGPAMSAFNAWVLLKGLETLAIRVQHSNASA QRIAEFLNGHPSVRWVRYPYLPSHPQYDLAKRQMSGGGTVVTFALDCPEDVAKQRAFEVLDKMRLIDISN NLGDAKSLVTHPATTTHRAMGPEGRAAIGLGDGVVRISVGLEDTDDLIADIDRALS
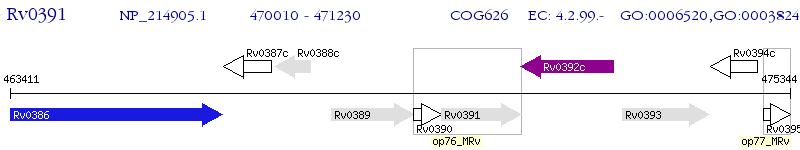
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0391 | metZ | - | 100% (406) | O-succinylhomoserine sulfhydrylase |
| M. tuberculosis H37Rv | Rv3340 | metC | 2e-63 | 35.53% (394) | O-acetylhomoserine aminocarboxypropyltransferase |
| M. tuberculosis H37Rv | Rv1079 | metB | 8e-63 | 37.05% (359) | cystathionine gamma-synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0397 | metZ | 0.0 | 100.00% (406) | O-succinylhomoserine sulfhydrylase |
| M. gilvum PYR-GCK | Mflv_0219 | - | 0.0 | 84.25% (400) | O-succinylhomoserine sulfhydrylase |
| M. leprae Br4923 | MLBr_00275 | metZ | 0.0 | 87.93% (406) | O-succinylhomoserine sulfhydrylase |
| M. abscessus ATCC 19977 | MAB_4242c | - | 0.0 | 82.54% (401) | O-succinylhomoserine sulfhydrylase |
| M. marinum M | MMAR_0688 | metZ | 0.0 | 87.10% (411) | O-succinylhomoserine sulfhydrylase, MetZ |
| M. avium 104 | MAV_4773 | metZ | 0.0 | 91.54% (402) | O-succinylhomoserine sulfhydrylase |
| M. smegmatis MC2 155 | MSMEG_0769 | metZ | 0.0 | 85.50% (407) | O-succinylhomoserine sulfhydrylase |
| M. thermoresistible (build 8) | TH_3089 | - | 0.0 | 87.28% (401) | PUTATIVE O-succinylhomoserine sulfhydrylase |
| M. ulcerans Agy99 | MUL_0201 | metB | 4e-61 | 35.93% (359) | cystathionine gamma-synthase |
| M. vanbaalenii PYR-1 | Mvan_0688 | - | 0.0 | 83.66% (410) | O-succinylhomoserine sulfhydrylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0219|M.gilvum_PYR-GCK --MSPEPDSVPSV------RIPTPLPDGVGQATIGVRGGLLRSGFEETAE
Mvan_0688|M.vanbaalenii_PYR-1 --MT----EVPSV------RIPAVLPEGVSQETIGVRGGLLRSGFEETAE
MSMEG_0769|M.smegmatis_MC2_155 --MT---DDIPSV------RIPAPLPDGVSQATIGVRGGLLRSGFEETAE
Rv0391|M.tuberculosis_H37Rv --MT----DESSV------RTPKALPDGVSQATVGVRGGMLRSGFEETAE
Mb0397|M.bovis_AF2122/97 --MT----DESSV------RTPKALPDGVSQATVGVRGGMLRSGFEETAE
MAV_4773|M.avium_104 MSQA----GDDSV------RTPPALPDGVSQATIGVRGGLLRSGFDETAE
MLBr_00275|M.leprae_Br4923 --MT----DDRSV------RTPKALPDGVTAATVGVRGGLLRSRFEETAE
MMAR_0688|M.marinum_M --MT----DDRSVPEAERPKNPRKLPDGVSQATIGVRGGMLRSGFDETAE
TH_3089|M.thermoresistible__bu --MT----DRPPASKPS-VRTPAGLPDGVGPATIGVRGGLLRSEFEETAE
MAB_4242c|M.abscessus_ATCC_199 --MTQTPSGGSSV------RIPKALPDGVSQATIGVRGGLLRSEFEETAE
MUL_0201|M.ulcerans_Agy99 ----------------------MKDDHKAQHRFAGLATRAIHSGYRPDPA
. . *: ::* : .
Mflv_0219|M.gilvum_PYR-GCK AIYLTSGYVYESAAEAEKAFTGEIDRYVYSRYGNPTISMFEERLRLLEGA
Mvan_0688|M.vanbaalenii_PYR-1 ALYLTSGYVYESAAAAEKAFTGEIDRYVYSRYGNPTVSMFEERLRLIEGA
MSMEG_0769|M.smegmatis_MC2_155 AMYLTSGYVYETAAAAEKAFTGDIDRYVYSRYGNPTISMFEERLRLIEGA
Rv0391|M.tuberculosis_H37Rv AMYLTSGYVYGSAAVAEKSFAGELDHYVYSRYGNPTVSVFEERLRLIEGA
Mb0397|M.bovis_AF2122/97 AMYLTSGYVYGSAAVAEKSFAGELDHYVYSRYGNPTVSVFEERLRLIEGA
MAV_4773|M.avium_104 ALYLTSGYVYESAAVAEQSFTGELDHFVYSRYGNPTVTMFEERLRLLEGA
MLBr_00275|M.leprae_Br4923 TMYLTSGYVYESAVVAEKSFTGELDHFVYSRYGNPTVTMFEERLRLIEGA
MMAR_0688|M.marinum_M ALYLTSGYVYESAAMAEQSFAGELDHFLYSRYGNPTVAMFEERLRLIEAA
TH_3089|M.thermoresistible__bu ALYLTSGYVYESAADAERAFAGEIDRFVYSRYGNPTIAMFEERLRLIEGA
MAB_4242c|M.abscessus_ATCC_199 AMYLTSGYVYESAAAAERAFTGEVDRYVYSRYGNPTISMFEERLRLIEGA
MUL_0201|M.ulcerans_Agy99 TGAVN-APIYASSTFAQDGVGGLRGGYEYARTGNPTRTALEAALAAVEDA
: :. . :* ::. *: .. * . : *:* **** : :* * :* *
Mflv_0219|M.gilvum_PYR-GCK PACFGTASGMSAVFTSLGALLGAGDRLVAARSLFGSCFVVCNEILPRWGV
Mvan_0688|M.vanbaalenii_PYR-1 PACFATSSGMSAVFTALGALLGAGDRLVAARSLFGSCFVVCNEILPRWGV
MSMEG_0769|M.smegmatis_MC2_155 PACFATATGMAAVFTALGALLGAGDRLVAARSLFGSCFVVCNEILPRWGV
Rv0391|M.tuberculosis_H37Rv PAAFATASGMAAVFTSLGALLGAGDRLVAARSLFGSCFVVCSEILPRWGV
Mb0397|M.bovis_AF2122/97 PAAFATASGMAAVFTSLGALLGAGDRLVAARSLFGSCFVVCSEILPRWGV
MAV_4773|M.avium_104 PAAFATASGMAAVFTSLGALLAAGDRLVAARSLFGSCFVVCNEILPRWGV
MLBr_00275|M.leprae_Br4923 PAAFVTASGMAAVFTSLGALLASGDRLVAARSLFGSCFVVCNEILPRWGV
MMAR_0688|M.marinum_M PAVFATASGMAAVFTALGALLGAGDRLVASRSLFGSCFVVCNEILPRWGV
TH_3089|M.thermoresistible__bu PACFATATGMAAVFTALGALLGAGDRLVAARSLFGSCFVVCNEILPRWGV
MAB_4242c|M.abscessus_ATCC_199 EAAFATATGMSAVFTALGALLGAGDRLVAARSLFGSCFVVCNEILPRWGV
MUL_0201|M.ulcerans_Agy99 AFGRAFSSGMAAADCALRAMLRPGDHVVIPDDAYGGTFRLIDKVFTGWNV
::**:*. :* *:* .**::* . . :*. * : .:::. *.*
Mflv_0219|M.gilvum_PYR-GCK ETVFVDGEDLAQWEEALSVPTQAVFFETPSNPMQSLVDIAAVCDLAHAAG
Mvan_0688|M.vanbaalenii_PYR-1 ETVFVDGDDLSQWEQALSVPTQAVFFETPSNPMQSLVDIAAVCELAHAAG
MSMEG_0769|M.smegmatis_MC2_155 ETVFVDGDDLSQWEEALSVPTTAVFFETPSNPMQSLVDIAAVSEMAHAAG
Rv0391|M.tuberculosis_H37Rv QTVFVDGDDLSQWERALSVPTQAVFFETPSNPMQSLVDIAAVTELAHAAG
Mb0397|M.bovis_AF2122/97 QTVFVDGDDLSQWERALSVPTQAVFFETPSNPMQSLVDIAAVTELAHAAG
MAV_4773|M.avium_104 QTVFVDGDDLAQWEEALSVPTAAVFFETPSNPMQSLVDIAAVTELAHAAG
MLBr_00275|M.leprae_Br4923 ETVFVDGEDLVQWEQALSVPTQAVFFETPSNPMQSLVDIAAVTELAHAAG
MMAR_0688|M.marinum_M ETVFVDGDDLEQWEQALSVPTEAVFFETPSNPMQSLVDIAAVTELAHAAG
TH_3089|M.thermoresistible__bu QTVFVDGDDLGQWERALSEPTTAVFFETPSNPMQSLVDIAAVTELAHAAG
MAB_4242c|M.abscessus_ATCC_199 ETVFVDGEDISQWEEALSVPTQAVFFETPSNPMQSLVDIEAVCTLAHASG
MUL_0201|M.ulcerans_Agy99 EYTPVALADLDAVRAAIRPTTRLIWVETPTNPLLSIADIAGIAQLGADSS
: . * *: . *: .* ::.***:**: *:.** .: :. :.
Mflv_0219|M.gilvum_PYR-GCK AKVVLDNVFATPILQQGMPLGADVVVYSGTKHIDGQGRVLGGAILGDKEY
Mvan_0688|M.vanbaalenii_PYR-1 AKVVLDNVFATPILQQGFPLGVDVVVYSGTKHIDGQGRVLGGAILGSKEY
MSMEG_0769|M.smegmatis_MC2_155 AKVVLDNVFATPLLQQGIPMGADVVVYSGTKHIDGQGRVLGGAILGDQEY
Rv0391|M.tuberculosis_H37Rv AKVVLDNVFATPLLQQGFPLGVDVVVYSGTKHIDGQGRVLGGAILGDREY
Mb0397|M.bovis_AF2122/97 AKVVLDNVFATPLLQQGFPLGVDVVVYSGTKHIDGQGRVLGGAILGDREY
MAV_4773|M.avium_104 AKVVLDNVFATPLLQQGIPLGVDVVVYSGTKHIDGQGRVLGGAILGDRDY
MLBr_00275|M.leprae_Br4923 AKVVLDNAFATPLLQHGLPLGVDVVIYSGTKHIDGQGRVLGGAILGDRDY
MMAR_0688|M.marinum_M AKVVLDNVFATPLLQQGIPLGVDVVVYSGTKHIDGQGRVLGGAILGDQDY
TH_3089|M.thermoresistible__bu AKVVLDNVFATPLLQQGFQLGVDVVVYSGTKHIDGQGRVLGGAILGDTDY
MAB_4242c|M.abscessus_ATCC_199 AKVVLDNVFATPLLQQGIPLGADVVVYSGTKHIDGQGRVLGGAILGETEY
MUL_0201|M.ulcerans_Agy99 AKVLVDNTFASPALQQPLSLGADVVLHSTTKYIGGHSDVVGGALVTNDEE
***::**.**:* **: : :*.***::* **:*.*:. *:***:: . :
Mflv_0219|M.gilvum_PYR-GCK IDGPVQKLMRHTGPALSPFNAWTLLKGLETMALRVQHQNSSAHRIAEFLE
Mvan_0688|M.vanbaalenii_PYR-1 IDEPVQKLMRHTGPALSPFNAWTLLKGLETLSLRVQHQNSSAHRIAEFLE
MSMEG_0769|M.smegmatis_MC2_155 IDGPVQKLMRHTGPAISAFNAWVLLKGLETLAVRVDYSNRSAQRVAEFLE
Rv0391|M.tuberculosis_H37Rv IDGPVQKLMRHTGPAMSAFNAWVLLKGLETLAIRVQHSNASAQRIAEFLN
Mb0397|M.bovis_AF2122/97 IDGPVQKLMRHTGPAMSAFNAWVLLKGLETLAIRVQHSNASAQRIAEFLN
MAV_4773|M.avium_104 IDGPVQKLMRHTGPAMSAFNAWVLLKGLETMAIRVEHSNSSAHRIAEFLE
MLBr_00275|M.leprae_Br4923 IDGPVQKLMRHTGPAMSAFNAWILLKGLETLAIRVDHCNSSAHRIAEFLE
MMAR_0688|M.marinum_M IDGPVQKLMRHTGPALSAFNAWILLKGLETLAIRVQHSNSSALRIAEFLQ
TH_3089|M.thermoresistible__bu IDGPVQKLMRHTGPAISAFNAWIMLKGLETLAVRVNHSNASAHRIAEFLE
MAB_4242c|M.abscessus_ATCC_199 IEGPVKTLMRHTGPALSPFNAWTLVKGLETLDLRVRHANDSAYKIAQFLE
MUL_0201|M.ulcerans_Agy99 LDQSFAFLQNGAGAVPGPFDAYLTMRGLKTLVLRMQRHSENAAAVAEFLA
:: .. * . :*.. ..*:*: ::**:*: :*: . .* :*:**
Mflv_0219|M.gilvum_PYR-GCK GHSAVSWVKYPFLESHPQYDLAKRQMTGGGTVVTFELAGA--------TK
Mvan_0688|M.vanbaalenii_PYR-1 QHPSVSWVKYPFLESHPQYDLAKRQMTGGGTVVTFELAGAGGAGNPDAAK
MSMEG_0769|M.smegmatis_MC2_155 GHPAVRWVKYPFLQSHPQYELAKRQMRGGGTVVTFELDGDD-------GK
Rv0391|M.tuberculosis_H37Rv GHPSVRWVRYPYLPSHPQYDLAKRQMSGGGTVVTFALDCPE-----DVAK
Mb0397|M.bovis_AF2122/97 GHPSVRWVRYPYLPSHPQYDLAKRQMSGGGTVVTFALDCPE-----DVAK
MAV_4773|M.avium_104 THPAVRWVRYPYLPSHPQYDLAKRQMSGGGTVITFALDCPD-----DKAK
MLBr_00275|M.leprae_Br4923 KHPAVSWVRYPFLVSHPQYDLAKRQMSGGGTVVTFALNSPE-----DAAK
MMAR_0688|M.marinum_M GHPAVRWVRYPYLPSHPQYDLAKRQMSGGGTVVTFALDAPE-----SAAK
TH_3089|M.thermoresistible__bu GHPAVRWVRYPFLESHPQYDLAKRQMRGGGTVITFELDAPP-----DGGK
MAB_4242c|M.abscessus_ATCC_199 QHPAVRWVRYPFLGTHPQYELAKRQMRGGGTVVTFELDADG-----DAGK
MUL_0201|M.ulcerans_Agy99 EHPAISTVLYPGLPSHPGHAVAARQMRGFGGMVSVRMRAGR---------
*.:: * ** * :** : :* *** * * :::. :
Mflv_0219|M.gilvum_PYR-GCK ERAFEVLDKLQIVDISNNLGDAKSLITHPATTTHRAMGPEGRAAIGLGDG
Mvan_0688|M.vanbaalenii_PYR-1 ERAFEVLDKLQIVDISNNLGDAKSLITHPATTTHRAMGPEGRAAIGLGDG
MSMEG_0769|M.smegmatis_MC2_155 ARAFEVLDKLRVIDISNNLGDAKSLITHPATTTHRAMGPEGRAAIGLGDG
Rv0391|M.tuberculosis_H37Rv QRAFEVLDKMRLIDISNNLGDAKSLVTHPATTTHRAMGPEGRAAIGLGDG
Mb0397|M.bovis_AF2122/97 QRAFEVLDKMRLIDISNNLGDAKSLVTHPATTTHRAMGPEGRAAIGLGDG
MAV_4773|M.avium_104 QRAFEVLDKLTLIDISNNLGDAKSLVTHPATTTHRAMGPEGRAAIGLGDG
MLBr_00275|M.leprae_Br4923 QRAFEVLDRLRLIDISNNFGDVKSLITHPATTTHRAMGPEGRAAIGLGDG
MMAR_0688|M.marinum_M RRAFEVLDKLRLIDISNNLGDTKSLITHPATTTHRAMGPEGRAAIGLGDA
TH_3089|M.thermoresistible__bu QRAFEVLDKLRLIDISNNLGDAKSLITHPATTTHRAMGAEGRAAIGLGDA
MAB_4242c|M.abscessus_ATCC_199 QRAFQVLDGTSLIDISNNLGDSKSLITHPATTTHRAMGPEGRAAIGLSDG
MUL_0201|M.ulcerans_Agy99 TAAEQLCAKTNIFILAESLGSVESLIEHPSAMTHASTAG---SQLEVPDD
* :: :. :::.:*. :**: **:: ** : . : : : *
Mflv_0219|M.gilvum_PYR-GCK VVRISVGLEGTEDLIADLDRALG
Mvan_0688|M.vanbaalenii_PYR-1 VVRISIGLEGTEDLLADLDRALG
MSMEG_0769|M.smegmatis_MC2_155 VVRISIGLEGTEDLIADLDQALS
Rv0391|M.tuberculosis_H37Rv VVRISVGLEDTDDLIADIDRALS
Mb0397|M.bovis_AF2122/97 VVRISVGLEDTDDLIADIDRALS
MAV_4773|M.avium_104 VVRISVGLEGTDDLIADIDRALG
MLBr_00275|M.leprae_Br4923 VVRISIGLEDAADLIADIDQALS
MMAR_0688|M.marinum_M VVRISVGLEGAEDLIADIDQALG
TH_3089|M.thermoresistible__bu VVRISVGLEDTEDLIADLDRALS
MAB_4242c|M.abscessus_ATCC_199 VVRLSVGLESTDDLIADLERALG
MUL_0201|M.ulcerans_Agy99 LVRLSVGIEDVADLLDDLKQALG
:**:*:*:*.. **: *:.:**.