For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TATQITGVVLAAGRSNRLGTPKQLLPYRDTTVLGATLDVARQAGFDQLILTLGGAASAVRAAMALDGTDV VVVEDVERGCAASLRVALARVHPRATGIVLMLGDQPQVAPATLRRIIDVGPATEIMVCRYADGVGHPFWF SRTVFGELARLHGDKGVWKLVHSGRHPVRELAVDGCVPLDVDTWDDYRRLLESVPS*
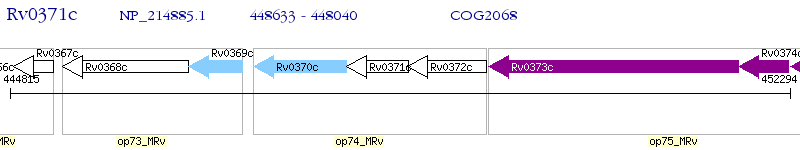
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0371c | - | - | 100% (197) | hypothetical protein Rv0371c |
| M. tuberculosis H37Rv | Rv0345 | - | 1e-05 | 30.70% (114) | hypothetical protein Rv0345 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0378c | - | 1e-111 | 100.00% (197) | hypothetical protein Mb0378c |
| M. gilvum PYR-GCK | Mflv_4787 | - | 4e-10 | 30.22% (182) | hypothetical protein Mflv_4787 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2937c | - | 1e-08 | 30.43% (161) | hypothetical protein MAB_2937c |
| M. marinum M | MMAR_0659 | - | 3e-80 | 73.71% (194) | hypothetical protein MMAR_0659 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1806 | - | 2e-08 | 30.52% (154) | hypothetical protein MSMEG_1806 |
| M. thermoresistible (build 8) | TH_0242 | - | 1e-11 | 29.57% (186) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0113 | - | 9e-79 | 73.20% (194) | hypothetical protein MUL_0113 |
| M. vanbaalenii PYR-1 | Mvan_1662 | - | 2e-08 | 33.12% (154) | hypothetical protein Mvan_1662 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1806|M.smegmatis_MC2_155 ---MSEPPVVAGVVLAAGAGTRFGMPKVLA--AEGDWLKTAVRSLVEGGC
TH_0242|M.thermoresistible__bu -MGVTTTSTAAGVVLAAGAGTRFGMPKILA--AEGLWLKIAVSALSGGGC
Mflv_4787|M.gilvum_PYR-GCK ---------MTGVVLAAGAGTRFGMPKVLA--DGGEWLRRSVAAVSDGGC
Mvan_1662|M.vanbaalenii_PYR-1 ----MTQELSTGVVLAAGAGTRFGMPKVLG--GDGDWLRRSVAALHDGGC
MAB_2937c|M.abscessus_ATCC_199 MEPDPTAGVVVGAVLAAGAGNRYGMPKILA--ANGRWLELAVTALDRGGC
Rv0371c|M.tuberculosis_H37Rv ----MTATQITGVVLAAGRSNRLGTPKQLLPYRDTTVLGATLDVARQAGF
Mb0378c|M.bovis_AF2122/97 ----MTATQITGVVLAAGRSNRLGTPKQLLPYRDTTVLGATLDVARQAGF
MMAR_0659|M.marinum_M ----MRAPSITGVVLAAGGSRRLGTPKQLLAYRDTTLLGATLRVARGAGF
MUL_0113|M.ulcerans_Agy99 ----MRAPSITGVVLAAGGSRRLGTPKQLLSYRDTTLLGATLRVARGAGF
.*.***** . * * ** * * :: .*
MSMEG_1806|M.smegmatis_MC2_155 AHVVVVLGAAVVDVPQP-----AQAVVATDWSDGLSASLRAGVGAAAR-T
TH_0242|M.thermoresistible__bu GDVVVVLGAAIVDVPAP-----ARAVVAQDWSRGLSASLRAGITALG--D
Mflv_4787|M.gilvum_PYR-GCK DDVVVVLGAAVVDVPAP-----ARAVVADDWRDGLSASVRAGVRAAS---
Mvan_1662|M.vanbaalenii_PYR-1 GDVVVVLGAAIVDVPAP-----ARAVVTADWADGLSASVRAGVRAAG--D
MAB_2937c|M.abscessus_ATCC_199 EEVYVTQGAVSVEMPSP-----AHGIEVARWRDGVSESVRAVLELVHART
Rv0371c|M.tuberculosis_H37Rv DQLILTLGGAASAVRAAMALDGTDVVVVEDVERGCAASLRVALARVHP--
Mb0378c|M.bovis_AF2122/97 DQLILTLGGAASAVRAAMALDGTDVVVVEDVERGCAASLRVALARVHP--
MMAR_0659|M.marinum_M DQLIVTLGGAAQRVRALVPLDEAEVVTVEDFGTGCSASLRVALGRVDP--
MUL_0113|M.ulcerans_Agy99 DQLIVTLGGAAQRVRALVPLDEAEVVTVEDFGAGCSASLRVALGRVDP--
.: :. *.. : : : . * : *:*. :
MSMEG_1806|M.smegmatis_MC2_155 GADLIVFRLVDTPDIGGDVIARVAAAAG--ESGLARATYDGRPGHPVVIA
TH_0242|M.thermoresistible__bu DADLAVIHTVDTPDIGADVVRRVLTAARSTPSGLARAWFGDRPGHPVVVG
Mflv_4787|M.gilvum_PYR-GCK GADFVVLTTVDTPDVGAAAVRRVLTAARESASGLVRARYGGRPGHPVVIA
Mvan_1662|M.vanbaalenii_PYR-1 AADFVVLTTVDTPDVGAAAVRRVLAAARASTSGLARAYYDGRPGHPVVIA
MAB_2937c|M.abscessus_ATCC_199 DVVGVLIHLVDLPSVGPEVVRMVLRASGGRRSALVRATFAGRPGHPVYLG
Rv0371c|M.tuberculosis_H37Rv RATGIVLMLGDQPQVAPATLRRIIDVGP--ATEIMVCRYADGVGHPFWFS
Mb0378c|M.bovis_AF2122/97 RATGIVLMLGDQPQVAPATLRRIIDVGP--ATEIMVCRYADGVGHPFWFS
MMAR_0659|M.marinum_M QSAGIVLMLGDQPGVDPATVARLVGQGP--GCDLMVCRYNDGIGHPFWFS
MUL_0113|M.ulcerans_Agy99 QSAGIVLMLGDQPGVDPATVARLVGQGP--GCDLMVCRYNDGIGHPFWFS
:: * * : .: : . : . : . ***. ..
MSMEG_1806|M.smegmatis_MC2_155 RRHWPELLRVVHGDEGAKPFLRGRTDVVAVECGDLATGADIDVSP-----
TH_0242|M.thermoresistible__bu RQHWPALLRSLHGDEGAKRFLRGRSDVVRVDCSDLAGGRDIDTPNSGADA
Mflv_4787|M.gilvum_PYR-GCK RRHLPQLLDSLSGDDGAGPFLKARADVVAVECGDLSSGRDIDTR------
Mvan_1662|M.vanbaalenii_PYR-1 RRHWPALLDALAGDVGAGPFLAGRDDVVAVECADLGTGRDIDAR------
MAB_2937c|M.abscessus_ATCC_199 SEHVGPVLSALAADRGAGPYLAARTDVIEVPCEHLADGADHDYRP-----
Rv0371c|M.tuberculosis_H37Rv RTVFGELAR-LHGDKGVWKLVHSGRHPVRELAVDGCVPLDVDTWDDYRRL
Mb0378c|M.bovis_AF2122/97 RTVFGELAR-LHGDKGVWKLVHSGRHPVRELAVDGCVPLDVDTWDDYRRL
MMAR_0659|M.marinum_M RRLFGELTQ-LHGDKGVWKLIQSGRHPVRELVVDDSVPLDVDTWDDYRRL
MUL_0113|M.ulcerans_Agy99 RRLFGELTQ-LHGDKGVWKLIQSGRHPVRELVVDDSVPLDVDTRDDYRRL
: : .* *. : . . : . * *
MSMEG_1806|M.smegmatis_MC2_155 ------
TH_0242|M.thermoresistible__bu R-----
Mflv_4787|M.gilvum_PYR-GCK ------
Mvan_1662|M.vanbaalenii_PYR-1 ------
MAB_2937c|M.abscessus_ATCC_199 ------
Rv0371c|M.tuberculosis_H37Rv LESVPS
Mb0378c|M.bovis_AF2122/97 LESVPS
MMAR_0659|M.marinum_M VESVP-
MUL_0113|M.ulcerans_Agy99 VESVP-