For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRWKQGWTRGSLFAALNIAAVVAVLMLGAGVAVADPDAAPGDPGGPGAPGAQRDPSTRRQLTCWRRHPT RWRCRRHLTRWRRRHLTRSRRPRLTRWQCR
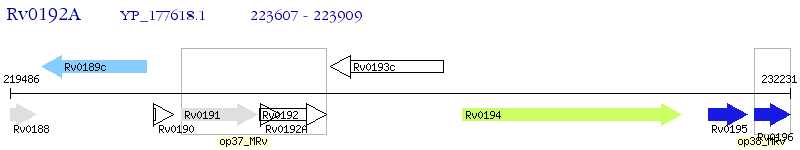
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0192A | - | - | 100% (100) | CONSERVED SECRETED PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0198 | - | 1e-25 | 91.07% (56) | hypothetical protein Mb0198 |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0435 | - | 2e-13 | 75.00% (44) | secretory protein |
| M. avium 104 | MAV_4986 | - | 7e-11 | 58.93% (56) | ErfK/YbiS/YcfS/YnhG family protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1085 | - | 1e-13 | 75.00% (44) | secretory protein |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0435|M.marinum_M MSRWTRGRIRGSLFAAVNAAGVVGV---LLLGAGPAVADPDDAPGDP-PV
MUL_1085|M.ulcerans_Agy99 MSRWMRGRIRGSLFAAVNAAGVVGV---LLLGAGPAVADPDDAPGDP-PV
MAV_4986|M.avium_104 MSGWTR----GTLFAALNAAVVSVVGLALVLSAGPALADPDPAPADPGAV
Mb0198|M.bovis_AF2122/97 MSRWKQGWTRGSLFAALNIAAVVAV---LMLGAGVAVADPDAAPGDPGGP
Rv0192A|M.tuberculosis_H37Rv MSRWKQGWTRGSLFAALNIAAVVAV---LMLGAGVAVADPDAAPGDP---
** * : *:****:* * * * *:*.** *:**** **.**
MMAR_0435|M.marinum_M IAPAEP--APDP-----LAPPPGPLALPPMPDPLAPPP---FPDPLAPPP
MUL_1085|M.ulcerans_Agy99 IAPAEP--APDP-----LAPPPGPLALPPMPDPLAPP------------P
MAV_4986|M.avium_104 AAPPGPPAPPDP-----LAPPPPPDPLAPPP-PAAPP-------APWLPP
Mb0198|M.bovis_AF2122/97 GGPGGTAGPVDPPAVDLLAPPPDPLALPPALDPLAPPPPDPLAPPPPDPL
Rv0192A|M.tuberculosis_H37Rv ---GGP--------------------------------------------
.
MMAR_0435|M.marinum_M LVPVAAGPVAGQDPTPFFGPPPFRPPSFNPVDGAMVGVAKPIIINFQVPI
MUL_1085|M.ulcerans_Agy99 LVPVAAGPVAGQDPTPFFGPPPFRPPSFNPVDGAMVGVAKPIIINFQVPI
MAV_4986|M.avium_104 AAQPAAAPAAGQDPTPFTGTPPFGPPTFVPKTGSTVGVAQPIIINFPGRV
Mb0198|M.bovis_AF2122/97 AVPVAAGPVAGQDPTPFVGPPPFRPPTFNPVDGAMVGVAKPIVINFAVPI
Rv0192A|M.tuberculosis_H37Rv -----GAPGAQRDPS-----------------------------------
..* * :**:
MMAR_0435|M.marinum_M ADQAMAESAIHISSIPPVPGKFYWMTPTQVRWRPFQFWPANTAVNIDAAG
MUL_1085|M.ulcerans_Agy99 ADQAMAESAIHISSIPPVPGKFYWMTPTQVRWRPFQFWPANTAVNIDAAG
MAV_4986|M.avium_104 DDAGAAISAVHVSSVPPVPGKFYWMTPTQLRWRPLSFWPAHTAVTVDAGG
Mb0198|M.bovis_AF2122/97 ADRAMAESAIHISSIPPVPGKFYWMSPTQVRWRPFEFWPANTAVNIDAAG
Rv0192A|M.tuberculosis_H37Rv -------TRRQLTCWRRHP----------TRWR-----------------
: :::. * ***
MMAR_0435|M.marinum_M TKSSFRTGDSLVATADDATHQMTITRNGVVEQTFPMSMGMAAGNHQTPNG
MUL_1085|M.ulcerans_Agy99 TKSSFRTGDSLVATADDATHQMTITRNGVVEQTFPMSMGMAAGNHQTPNG
MAV_4986|M.avium_104 TVTNFQTGDTLVATADDATHQLTVTRNGTVEKTFPMSMGMTAGNHQTPNG
Mb0198|M.bovis_AF2122/97 TKSSFRTGDSLVATADDATHQMTITRNGVVQKTFPMSMGMVSGGHQTPNG
Rv0192A|M.tuberculosis_H37Rv -----------------CRRHLTRWRR----------------RHLTRS-
. :::* *. * * .
MMAR_0435|M.marinum_M TYYVLEKMPTVVMDSSTYGVPVNSPQGYKVTVADAVRIDNSGNFVHSAPW
MUL_1085|M.ulcerans_Agy99 TYYVLEKMPTVVMDSSTYGVPVNSPQGYKVTVADAVRIDNSGNFVHSAPW
MAV_4986|M.avium_104 TYYVQDKKASVVMDSSTYGVPVNSTYGYKVTVEDAVRFDNVGDYVHSAPW
Mb0198|M.bovis_AF2122/97 TYYVLEKFATVVMDSSTYGVPVNSAQGYKLTVSDAVRIDNSGNFVHSAPW
Rv0192A|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0435|M.marinum_M SVGDQGKRDVSHGCINLSPTNAKWFFDNFGSGDPIVVKNSVG-TYNKNDG
MUL_1085|M.ulcerans_Agy99 SVGDQGKRDVSHGCINLSPTNAKWFFDNFGSGDPIVVKNSVG-TYNKNDG
MAV_4986|M.avium_104 SVDDQGKRDVSHGCINISPANAKWFFDNFGPGDPIIVKNSSGGDYKKNDG
Mb0198|M.bovis_AF2122/97 SVADQGKRNVTHGCINLSPANAKWFYDNFGSGDPVVVKNSVG-TYNKNDG
Rv0192A|M.tuberculosis_H37Rv ------RR----------PRLTRWQCR-----------------------
:* * ::*
MMAR_0435|M.marinum_M AQDWQI
MUL_1085|M.ulcerans_Agy99 AQDWQI
MAV_4986|M.avium_104 SADWMN
Mb0198|M.bovis_AF2122/97 AQDWQI
Rv0192A|M.tuberculosis_H37Rv ------