For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTPVTTFPLVDAILAGRDRNLDGVILIAAQHLLQTTHAMLRSLFRVGLDPRNVAVIGKCYSTHPGVVDAM RADGIYVDDCSDAYAPHESFDTQYTRHVERFFAESWARLTAGRTARVVLLDDGGSLLAVAGAMLDASADV IGIEQTSAGYAKIVGCALGFPVINIARSSAKLLYESPIIAARVTQTAFERTAGIDSSAAILITGAGAIGT ALADVLRPLHDRVDVYDTRSGCMTPIDLPNAIGGYDVIIGATGATSVPASMHELLRPGVLLMSASSSDRE FDAVALRRRTTPNPDCHADLRVADGSVDATLLNSGFPVNFDGSPMCGDASMALTMALLAAAVLYASVAVA DEMSSDHPHLGLIDQGDIVASFLNIDVPLQALSRLPLLSIDGYRRLQVRSGYTLFRQGERADHFFVIESG ELEALVDGKVILRLGAGDHFGEACLLGGMRRIATVRACEPSVLWELDGKAFGDALHGDAAMREIAYGVAR TRLMHAGASESLMV
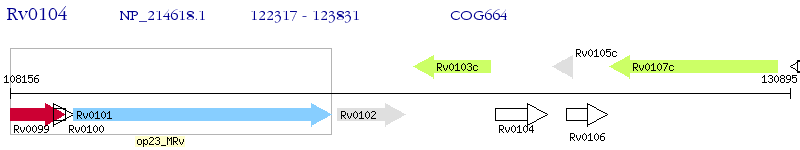
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0104 | - | - | 100% (504) | hypothetical protein Rv0104 |
| M. tuberculosis H37Rv | Rv2565 | - | 9e-08 | 32.93% (82) | hypothetical protein Rv2565 |
| M. tuberculosis H37Rv | Rv3239c | - | 8e-07 | 28.12% (192) | transmembrane transport protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0107 | - | 0.0 | 99.60% (504) | hypothetical protein Mb0107 |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00187 | - | 5e-05 | 35.71% (98) | hypothetical protein MLBr_00187 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0285 | - | 0.0 | 73.66% (505) | hypothetical protein MMAR_0285 |
| M. avium 104 | MAV_1121 | - | 5e-06 | 37.93% (87) | cyclic nucleotide-binding protein |
| M. smegmatis MC2 155 | MSMEG_5458 | - | 2e-07 | 40.23% (87) | cyclic nucleotide-binding protein |
| M. thermoresistible (build 8) | TH_0935 | - | 8e-07 | 37.50% (104) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4667 | - | 0.0001 | 37.65% (85) | hypothetical protein MUL_4667 |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5458|M.smegmatis_MC2_155 --------------------------------------------------
TH_0935|M.thermoresistible__bu --------------------------------------------------
MAV_1121|M.avium_104 --------------------------------------------------
MUL_4667|M.ulcerans_Agy99 -------------------------------------------------M
MLBr_00187|M.leprae_Br4923 -------------------------------------------------M
Rv0104|M.tuberculosis_H37Rv MTPVTTFPLVDAILAGRDRNLDGVILIAAQHLLQTTHAMLRSLFRVGLDP
Mb0107|M.bovis_AF2122/97 MTPVTTFPLVDAILAGRDRNLDGVILIAAQHLLQTTHAMLRSLFRVGLDP
MMAR_0285|M.marinum_M MTQVTTFPLVDAIVSRRGGSLDGVVVIAAQHLLETTHAMLRSLFRVGLDP
MSMEG_5458|M.smegmatis_MC2_155 ----------------------MAELTEVRAADLAALEFFTGCRPSALEP
TH_0935|M.thermoresistible__bu ----------------------MTKVTTANAENLAGLELFAGYDGEALRP
MAV_1121|M.avium_104 ----------------------MAGLTGARADELATMDIFAGCPAEDLAP
MUL_4667|M.ulcerans_Agy99 DG--------------------MAEVTGAWVGELAGMDIFAGCAAQDLMP
MLBr_00187|M.leprae_Br4923 AGSWQCGHCESCASPLGPRDIAVVELIADRAEEFAAMDIFRGLPAEDLMS
Rv0104|M.tuberculosis_H37Rv RNVAVIGKCYSTHPGVVDAMRADGIYVDDCSDAYAPHESFDTQYTRHVER
Mb0107|M.bovis_AF2122/97 RNVAVIGKCYSTHPGVVDAMRADGIYVDDCSDAYAPHESFDTQYTRHVEW
MMAR_0285|M.marinum_M RNVAVIGKCYSTHLGVADAMRADGVHVDEFSAAYAPHESFDTEYTRHVER
* : * :
MSMEG_5458|M.smegmatis_MC2_155 LATQLRPLKAE---PGQVLIRQGD---PALTFMLIESGRVQVSHAVADGP
TH_0935|M.thermoresistible__bu LAALLRPLTAE---PGTVLMHQGE---PAVHFLLIGSGRVEITHTGPDGA
MAV_1121|M.avium_104 LARRLRPLRAP---AGQVLMRQGE---QAVEFLLISSGSAEVKHVGDDGV
MUL_4667|M.ulcerans_Agy99 LAARLAPLRAA---AGQILMRQGE---QAVSFLLISSGSAEVIHVGDDGV
MLBr_00187|M.leprae_Br4923 VAVSVEPVLAA---AGEVLMQQGE---QAVSFLLISSGNVEVRRVDDDGA
Rv0104|M.tuberculosis_H37Rv FFAESWARLTAGRTARVVLLDDGGSLLAVAGAMLDASADVIGIEQTSAGY
Mb0107|M.bovis_AF2122/97 FFAESWARLTAGRTARVVLLDDGGSLLAVAGAMLDASADVIGIEQTSAGY
MMAR_0285|M.marinum_M FFAQSWDRLAAGRAGRVVLLDDGGSLLAAAGAALDGTANIVGIEQTSAGY
. : :*: :* . * :. . *
MSMEG_5458|M.smegmatis_MC2_155 PIVLDIEPGLIIGEIA-----LLRDAPRTATVVAAEPVIGWVGDRDAFDT
TH_0935|M.thermoresistible__bu VVVTEVGPGVIVGEIS-----LLRNAPRTATVVAAEPLSGWIGDREAFSV
MAV_1121|M.avium_104 VITEHTSPGMIVGEIA-----LLRDIPRTATVTTAQPLTGWTGDTEAFAS
MUL_4667|M.ulcerans_Agy99 VIVEQALPGMIVGEIA-----LLRNVPRTATVTTVEPLTGWIGGTDAFDL
MLBr_00187|M.leprae_Br4923 VIVGQASHGMIIGEIA-----LLRDGRRTATVITTEPLTGWVGDIDAFAQ
Rv0104|M.tuberculosis_H37Rv AKIVGCALGFPVINIARSSAKLLYESPIIAARVTQTAFERTAGIDSSAAI
Mb0107|M.bovis_AF2122/97 AKIVGCALGFPVINIARSSAKLLYESPIIAARVTQTAFERTAGIDSSAAI
MMAR_0285|M.marinum_M AKIANCALGFPVVNIARSSAKLLYESPIIAGNVTRCAFDRIEGLDRGDAI
*. : :*: ** : * : .. * .
MSMEG_5458|M.smegmatis_MC2_155 ILH----LPGMFDRLVRIARQRLAAFIT------PIPVQVRTGEWFYLR-
TH_0935|M.thermoresistible__bu MLG----IPGFADTLVKIARQRLASFIT------PIPVRMRDRTRLFLR-
MAV_1121|M.avium_104 MVH----IPGVMPRLLRTVRQRLAAFIT------PIPIQVRDGTHLLLR-
MUL_4667|M.ulcerans_Agy99 MVH----IPGIMDRLVRTVRQRLAAFIT------PIPVPLRDGTQLMLR-
MLBr_00187|M.leprae_Br4923 MVQ----IPSITRRLLLTVRQRLAAFIT------PIPVQLRDGTHLMLR-
Rv0104|M.tuberculosis_H37Rv LITGAGAIGTALADVLRPLHDRVDVYDTRSGCMTPIDLPNAIGGYDVIIG
Mb0107|M.bovis_AF2122/97 LITGAGAIGTALADVLRPLHDRVDVYDTRSGCMTPIDLPNAIGGYDVIIG
MMAR_0285|M.marinum_M LITGAGAIGSALADQLRPRHERVDVYDTRVGHPGPIDLTQAIGDYDVIIG
:: : : ::*: : * ** : :
MSMEG_5458|M.smegmatis_MC2_155 ----PVLPGDVERTLNG----------PVEFSSETLYRRFQS-------V
TH_0935|M.thermoresistible__bu ----PVLPGDEAKTREG----------HVEFSNETLYRRFMS-------M
MAV_1121|M.avium_104 ----PVLPGDSERTVHG----------HIHFSSDTLYRRFMS-------P
MUL_4667|M.ulcerans_Agy99 ----PVLPGDSERTVHG----------HVHFSSETIYRRFMS-------A
MLBr_00187|M.leprae_Br4923 ----PVLPGDTERSLRG----------HVRFSRETLYLRFMS-------A
Rv0104|M.tuberculosis_H37Rv ATGATSVPASMHELLRPGVLLMSASSSDREFDAVALRRRTTPNPDCHADL
Mb0107|M.bovis_AF2122/97 ATGATSVPASMHELLRPGVLLMSASSSDREFDAVALRRRTTPNPDCHADL
MMAR_0285|M.marinum_M ATGATSVPTELHDLLRPGVVLMSASSSDREFSGAALRRRAIPDPNCHADL
. :* . . .*. :: * .
MSMEG_5458|M.smegmatis_MC2_155 RKPTRALLEYLFEVDYADHFVWVMTEGALGPVIADARFVREGHNATMAEV
TH_0935|M.thermoresistible__bu GQPNSALMHYLFAVDYVDHFVWVLTDTPDGVVVADGRFVRDHADPGTAEV
MAV_1121|M.avium_104 RLPTPALMHYLSEVDYVDHFVWVVTDGAD--PVADARFVRDENDPTVAEI
MUL_4667|M.ulcerans_Agy99 RVPDLAMMRYLSEVDYVDHFVWVMVDGSD--PVADARFIRDSVDPAVAEI
MLBr_00187|M.leprae_Br4923 RAPSDELMHYLSEVDYVDHFVWVVTDGGD--PVADARFVRDESDPTLAEI
Rv0104|M.tuberculosis_H37Rv RVADGSVDATLLNSGFPVNFDGSPMCGDASMALTMALLAAAVLYASVAVA
Mb0107|M.bovis_AF2122/97 RVADGSVDATLLNSGFPVNFDGSPMCGDASMALTMALLAAAVLYASVAVA
MMAR_0285|M.marinum_M RIADGKLDATLLNSGFPVNFDGSAMCGDESMSLTMALLAAAVLYAASAEA
. : * .: :* :: . : . *
MSMEG_5458|M.smegmatis_MC2_155 AFTVGDD------YQGRGIGSFLMGALIVSAN-------YVGVQRFNAR-
TH_0935|M.thermoresistible__bu AFLVADD------YQGRGVGSFLMDALAVAAT-------CAGVQRFTAQ-
MAV_1121|M.avium_104 AFTVADA------YQGRGIGTFLISALSIAAE-------VDGIERFSAR-
MUL_4667|M.ulcerans_Agy99 AFTVADA------YQGKGIGSFLVGALAVAAL-------VDGVARFSAR-
MLBr_00187|M.leprae_Br4923 AFTVADA------YQGRGVGNFLISALSIAAH-------VNGVNRFSAR-
Rv0104|M.tuberculosis_H37Rv DEMSSDHPHLGLIDQGDIVASFLNIDVPLQALSRLPLLSIDGYRRLQVRS
Mb0107|M.bovis_AF2122/97 DEMSSDHPHLGLIDQGDIVASFLNIDVPLQALSRLPLLSIDGYRRLQVRS
MMAR_0285|M.marinum_M ADIDCDHPHLGLTDQGDIVQSFLNIDVPLQALGRLPLLSVGGYRRIELRS
* ** : .** : : * * *: :
MSMEG_5458|M.smegmatis_MC2_155 --------------VLTDNMAMRKIMD-----------RLGAVWVREDLG
TH_0935|M.thermoresistible__bu --------------VLVDNYPMRRILD-----------RFGVHWEFADNN
MAV_1121|M.avium_104 --------------MLSDNVPMRAIMD-----------RYGAVWQREDIG
MUL_4667|M.ulcerans_Agy99 --------------MLADNLPMRTIMD-----------RHGAVWQREDVG
MLBr_00187|M.leprae_Br4923 --------------MLTDNGPMRAIMD-----------HHGAVWRRYDVG
Rv0104|M.tuberculosis_H37Rv GYTLFRQGERADHFFVIESGELEALVDGKVILRLGAGDHFGEACLLGGMR
Mb0107|M.bovis_AF2122/97 GHTLFRQGERADHFFVIESGELEALVDGKVILRLGAGDHFGEACLLGGMR
MMAR_0285|M.marinum_M GDIVFRQGEPASHFYVVESGELEAFVDGQLVLRLVAGDHFGEIALLSGWR
: :. :. ::* : * .
MSMEG_5458|M.smegmatis_MC2_155 VVMTEVDVPPVDTVPFEPELIDQIRDATRKVIRAVSQ-------------
TH_0935|M.thermoresistible__bu VVSTVFDVPKLRDLSLSPRLYRRIYDMARQVVRAVG--------------
MAV_1121|M.avium_104 VITTVIDVPHRRDLPFRKHEADEIRRVARQVIETVG--------------
MUL_4667|M.ulcerans_Agy99 VITTVIDVPGPRDLGFGREMAGQIRRVARQVIEAVS--------------
MLBr_00187|M.leprae_Br4923 VITTVIDVPRQRDLIIGRAMADQIAGVVRQVIGAVG--------------
Rv0104|M.tuberculosis_H37Rv RIATVRACEPSVLWELDGKAFGDALHGDAAMREIAYGVARTRLMH-AGAS
Mb0107|M.bovis_AF2122/97 RIATVRACEPSVLWELDGKAFGDALHGDAAMREIAYGVARTRLMH-AGAS
MMAR_0285|M.marinum_M RAATVRASQPSVLWSLDAAAFSEVLRVDATMRELAHQAIHSRVSRVVGAP
* : : .
MSMEG_5458|M.smegmatis_MC2_155 -----
TH_0935|M.thermoresistible__bu -----
MAV_1121|M.avium_104 -----
MUL_4667|M.ulcerans_Agy99 -----
MLBr_00187|M.leprae_Br4923 -----
Rv0104|M.tuberculosis_H37Rv ESLMV
Mb0107|M.bovis_AF2122/97 ESLMV
MMAR_0285|M.marinum_M KSSSV