For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDGDFTPAQQLKFALLDEGLALHGSAWTALRELNGTRSLSPHDYASTSGVILELDDDVWVNAPIEAFNPN FVTQSPYQLLYDNGFRVEGQGLVSRATFWPQPAYHSTTGPWGPWNNFVVTHGDRGRLSPLRSCAMTCTFC NIPYDDAIDVYSLKPVDALLAAAHLAIDDPIQPARHLLISGGTPKPKDIGWMREFYERMLTEFPNIELDI MMVPLPGLFDLPRLAALGLHELSINIELFNRDLTREFARQKYNQGLALYLDFIEAATTELGVGRARSMLM VGLEPVEDTLAGVRAIAERGGTPVLSPFRPDPATPLRQLKPPSAETMIDTYLRARDITEEHGVALGPDCP PCSHNTLNFATDSAGALHYQRDRPRMLGAC
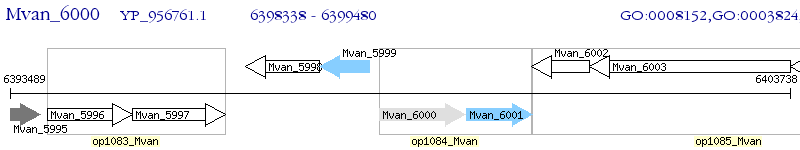
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_6000 | - | - | 100% (380) | radical SAM domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1221 | - | 3e-05 | 27.68% (112) | radical SAM domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0568 | - | 4e-05 | 23.97% (242) | radical SAM domain-containing protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_6000|M.vanbaalenii_PYR-1 MDGDFTPAQQLKFALLDEGLALHGSAWTALRELNGTRSLSPHDYASTSGV
MSMEG_0568|M.smegmatis_MC2_155 ---------------------MSTRVDLALLGFRGRPPVHRNAGAGPSAD
: . ** :.* .: : *..*.
Mvan_6000|M.vanbaalenii_PYR-1 ILELDDDVWVNAPIEAFNPNFVTQSPYQLLYDNGFRVEGQGLVSRATFWP
MSMEG_0568|M.smegmatis_MC2_155 GHLVIDGLNAAIPRNPLSP-FVFDGTRVLLDG-----EDTGLDVEVIDRP
: *.: . * :.:.* ** :.. ** . *. ** .. *
Mvan_6000|M.vanbaalenii_PYR-1 QPAYHSTTGPWGPWNNFVVTHGDRGRLSPLRSCAMT------CTFCNIPY
MSMEG_0568|M.smegmatis_MC2_155 R-FYDLTTGDGVPYEKLARLHGRNVLATTVVQTCIRYSPDQRCRFCTIEE
: *. *** *::::. ** . :.: . .: * **.*
Mvan_6000|M.vanbaalenii_PYR-1 DDAIDVYSLKPVDALLAAAHLAIDDPIQPARHLLISGGTPKPKDIG--WM
MSMEG_0568|M.smegmatis_MC2_155 SLRSGATTAVKRPAELAEVAAAAVR-LDGVTQMVMTTGTSAGTDRGARHL
. .. : * ** . * :: . ::::: **. .* * :
Mvan_6000|M.vanbaalenii_PYR-1 REFYERMLTEFPNIELDIMMVPLPGLFDLPRLAALGLHELSINIELFNRD
MSMEG_0568|M.smegmatis_MC2_155 ARCVRAVKAAVPALPIQVQCEPPADLAVLSELRAAGADAIGIHVESLDDE
. . : : .* : ::: * ..* *..* * * . :.*::* :: :
Mvan_6000|M.vanbaalenii_PYR-1 LTREFARQKYNQGLALYLDFIEAATTELGVGRARSMLMVGLEP-VEDTLA
MSMEG_0568|M.smegmatis_MC2_155 VRSRWMPGKATVALDAYRAAWREAVRVFGRNQVSTYLLVGLGENPDELVA
: .: * . .* * . *. :* .:. : *:*** :: :*
Mvan_6000|M.vanbaalenii_PYR-1 GVRAIAERGGTPVLSPFRPDP---ATPLRQLKPPSAETMIDTYLRARDIT
MSMEG_0568|M.smegmatis_MC2_155 GAGELIDMGVYPFVVPFRPQSGSLATDVDGAIAPDSGLVEKVSRAVADLL
*. : : * *.: ****:. ** : .*.: : .. . *:
Mvan_6000|M.vanbaalenii_PYR-1 EEHGVALGPDCPPCSHNTLNFATDSAGALHYQRDRPRMLGAC
MSMEG_0568|M.smegmatis_MC2_155 RQAGMSGADQRAGCAACGACSVLQNLGA--------------
.: *:: . : . *: . :. **