For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IDEHGHLHLRSAEHLDPIWRLWDELPAQWRGPVLGPGIQNWAAITENDWPHLDLSGLPDPFAAELAWMAH WQASDGTRVSVLAMAQLANMIRRAVVEGRDIPSSIRAMDYDAAAVLQSWFYLSRGKRLPSARGRGRVHAL FGFARHALIAASHDRPWWQLDFWHPRCDPRIPLTDREPVANYGCSPGDIQLPWVRAAVKWHLGTMLESGA LRWTTVSQERMRSFRRFDNWLTANFDDPTDILGDPTTAVEQAAAFARWTAVAANRVTRQSDTRHLGRTVP IRGINDDLRAVAGLLEFVAANPGEARSVLGAEPWQRVNAAHSSSWFGRVTRIPHQRGFNDANYIDDHALA QITAALPLIGLPRTEQMTITRGDGTTLTANGLDDPQAMRMILLQILTGRRASEIRTCDFDCLSAAPTSSG AAAGDDQSLTRFRYAQSKIDVAPDTILVDREVTEVIAEQHRWLQAQHPDGSRRYLFARRLGNRRGDKPYP AGTYNWVLRTLSDLVQITDAKGQVVRLSHTHRFRHTRLTRLAELGLPIHVLQRYAGHATPTMTMHYIAAR DEHAEQAFLATAKLRSDGTRIQLSSDDHDSLHLFRRADRFLPNGWCMLPPLQSCDKGNACLTCSVFVTDH THRDVLERQLRETSDLIDWTTTEFENRHSRPMPEDNVWLIRRHAEHHALTKLLDALNHAPDSATHGATTK CHRPSAGPVPVPLDLTRHRRRTR*
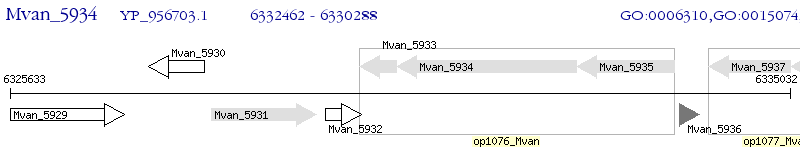
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5934 | - | - | 100% (724) | phage integrase family protein |
| M. vanbaalenii PYR-1 | Mvan_3903 | - | 0.0 | 100.00% (724) | phage integrase family protein |
| M. vanbaalenii PYR-1 | Mvan_3897 | - | 0.0 | 100.00% (724) | phage integrase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4644 | - | 3e-07 | 29.10% (189) | phage integrase family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0443 | - | 1e-83 | 31.28% (697) | putative transposase/integrase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0504 | - | 2e-88 | 34.06% (646) | putative transposase |
| M. smegmatis MC2 155 | MSMEG_4156 | - | 3e-21 | 32.71% (214) | putative transposase |
| M. thermoresistible (build 8) | TH_3646 | - | 2e-88 | 31.51% (733) | PUTATIVE phage integrase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0443|M.abscessus_ATCC_1997 MTPTNPPEDLTPKSAGPDTSWAHQWAQVPPRWRTPIYRIDTAPASEVFTL
TH_3646|M.thermoresistible__bu -------------VAGLG-SWEAQWAQVPAAWRRPVYPIYAAPYDEVFLR
MAV_0504|M.avium_104 --------------------------------------------------
Mvan_5934|M.vanbaalenii_PYR-1 --MIDEHGHLHLRSAEHLDPIWRLWDELPAQWRGPVLGPGIQNWAAITEN
MSMEG_4156|M.smegmatis_MC2_155 --------MTALDPNSTLGLLGRLMAGVRSEFRRDVLEFAADDAVFGGGS
Mflv_4644|M.gilvum_PYR-GCK -----------MLPEHDYAPTPETPAQIYAAYLVHLQRRDRG--------
MAB_0443|M.abscessus_ATCC_1997 NNYYRTRSNG-QGYDFTPAGPPRFADELAWWVYICWREGLCRIDPGDLKW
TH_3646|M.thermoresistible__bu NQFYLRANRATAAHDFTPAAPSRFAEEIAWWVWLCWDQQLRRIEPSLLAW
MAV_0504|M.avium_104 -------------------------------MWLCHHEGLRRIEPSVLKW
Mvan_5934|M.vanbaalenii_PYR-1 ----------DWPHLDLSGLPDPFAAELAWMAHWQASDGTR-----VSVL
MSMEG_4156|M.smegmatis_MC2_155 CR--------VTDCGRSARGHGLCQGHHQRWANAGRPDLEEFTVSTDPRW
Mflv_4644|M.gilvum_PYR-GCK ----------NTAYAQAARSFLRRWPRVQSWADIPLDEQLAANCSTRPFV
:
MAB_0443|M.abscessus_ATCC_1997 FLRALPAVIEDYQRRHGRPPHSLLDLTASEIMRH-----AVVSFERRNGR
TH_3646|M.thermoresistible__bu LVRTLPGAVAEHSARTGHPPVSIADLDPTELIRQ-----AALAFQRRNNR
MAV_0504|M.avium_104 AGQALTAAAADYRNTHGHQPTSITDLSAATIVRH-----AMLVFEKRNAR
Mvan_5934|M.vanbaalenii_PYR-1 AMAQLANMIRRAVVEGRDIPSSIRAMDYDAAAVL-----QSWFYLSRGKR
MSMEG_4156|M.smegmatis_MC2_155 RKHRPNQACRVDGCGYGSARGGLCQLHAQRWERAGRPHLASWLRDPLPVK
Mflv_4644|M.gilvum_PYR-GCK TFLMVSRRLQPGYDYLVHRKLSSLWHELTDSCLQP-------DLDQFISA
.
MAB_0443|M.abscessus_ATCC_1997 LPTAGYRRNLGYTIDKLFVLLAVRCSEAPWWSRDLWDLRIDPRIPQREHE
TH_3646|M.thermoresistible__bu LPSAGSRRNIRHLIEHLHLAVAVRCTDAPWWSHDIWDLRADPRIPQRPHE
MAV_0504|M.avium_104 LPSPGARRNVTHLIEHLHLYLSVRTTDTDWWAHDTWDLRADPRIPQREHE
Mvan_5934|M.vanbaalenii_PYR-1 LPSARGRGRVHALFGFARHALIAASHDRPWWQLDFWHPRCDPRIPLTDRE
MSMEG_4156|M.smegmatis_MC2_155 QPRPGATCRIRHCSLWPQAESPLCHSHTNTWKANGRTDIDGFVTRFESND
Mflv_4644|M.gilvum_PYR-GCK ALELGFTERVASAIGSQIIARLLIQTGRPLTGLRESDLQELLHACHVRQE
.: .:
MAB_0443|M.abscessus_ATCC_1997 PRHEQALKLAGIEPDWLREGVRFWMRTALTHQLFTWTTVISRSGNLGRQL
TH_3646|M.thermoresistible__bu PHHDQAVRLRGIEPVWLREGLRFWLRSALTYDLLTWSSVVSRARNIGSQL
MAV_0504|M.avium_104 PGHDQSVKIGAITPDWLREGVRFWLRTALDAELLRWSSAVERARDMARHF
Mvan_5934|M.vanbaalenii_PYR-1 PVANYGCSPGDIQLPWVRAAVKWHLGTMLESGALRWTTVSQERMRSFRRF
MSMEG_4156|M.smegmatis_MC2_155 TPANETIQLGMLTPQLKLEMQYVLQQRHDDRRGKLTPTVVARVVRLLIDT
Mflv_4644|M.gilvum_PYR-GCK RTGRGAKHYRSTTHSARQILFHLGILDTQARPAVTALTLEQRMVDVPVAL
: .
MAB_0443|M.abscessus_ATCC_1997 G-----TFCVARGHLHDPLISTDPDTLRDVFLDFLDHLRSPQASANKRG-
TH_3646|M.thermoresistible__bu G-----RFARDRGYLHDPLISTDPDELREAFLGYLEYLRSPQAATQSER-
MAV_0504|M.avium_104 G-----AFATEHGYA-DPALADDPAQLRLIFTAFTEYLRSPAAAATPDKP
Mvan_5934|M.vanbaalenii_PYR-1 DNWLTANFDDPTDILGDPTTAVEQAAAFARWTAVAANRVTRQSDTRHLGR
MSMEG_4156|M.smegmatis_MC2_155 ATTSLLDFDADEWRQRSAVLLNDTRSRGLLLYAHSTVLDLAEAGGWEAEY
Mflv_4644|M.gilvum_PYR-GCK R----PAFVAYLNRKYATCVPKTVSSLATRLAHFGRYLAAADPSLTSLNQ
* . .
MAB_0443|M.abscessus_ATCC_1997 LTDYM-VAEIQYEVQSFYTFMYDHAAEAAAATSNMRWKDITVGHTRLWA-
TH_3646|M.thermoresistible__bu LTPYT-VASLQSETQSFYTFMHDHAAEAAAGTGNLRWREATLTHTALWS-
MAV_0504|M.avium_104 LTPSA-IDATQSYTQVFYTFMVDHAPEAAAATGNPRWLALTDAHTRLWG-
Mvan_5934|M.vanbaalenii_PYR-1 TVPIRGINDDLRAVAGLLEFVAANPGEARSVLGAEPWQRVNAAHSSSWFG
MSMEG_4156|M.smegmatis_MC2_155 PRDVWRMHRLG--YEGHYTLRFDRIPQPWLREPAKRWIRLRLSRGLNLEA
Mflv_4644|M.gilvum_PYR-GCK LDRRRHIEPFITSLTTATNSVTGEPITVADRIRCIHAVGNFLAEITEWG-
: . .
MAB_0443|M.abscessus_ATCC_1997 ----------PAYQPRRAKKYRELTWYSTADLQRMLTYLDVLAARTSQRI
TH_3646|M.thermoresistible__bu ----------PIHAPRRQRSERDLTWYSTADLQRMLTYLDVLAADRKQKV
MAV_0504|M.avium_104 ----------PAFRARRANRVRELSWYSTGELQQMLAYLDVLAADRGTLV
Mvan_5934|M.vanbaalenii_PYR-1 ----------RVTRIPHQRGFNDANYIDDHALAQITAALPLIGLPRTEQM
MSMEG_4156|M.smegmatis_MC2_155 GGGRPLLAIARFGRFLAEVDIEDISRIDRELLERYLAHLNREYSPQRRGA
Mflv_4644|M.gilvum_PYR-GCK --------------WDDAPPRRLIFRTDLPRPPRCLPRYLPVDSDRKLTA
. . : .
MAB_0443|M.abscessus_ATCC_1997 ILNQPDGTITVAPG---LGDPQAARIWLLQAMTGRRASEILMLDHDPLSA
TH_3646|M.thermoresistible__bu VMTRPDGTLTVLPG---LGDPQAARIWLLQALTGRRASEILMLDYDPLEA
MAV_0504|M.avium_104 ALTHPDGTASIIKG---LGDPQAARIWLLQALTGRRASEILMLDHQPLQP
Mvan_5934|M.vanbaalenii_PYR-1 TITRGDGTTLTANG---LDDPQAMRMILLQILTGRRASEIRTCDFDCLSA
MSMEG_4156|M.smegmatis_MC2_155 HIGLLNGFFAAIRQHCWATGLPATAMFFAEDHPKRGEHLPRALAEHVMTQ
Mflv_4644|M.gilvum_PYR-GCK ALAKSP-------------YRLAADALLVQRACGLRIGELLDLELDCIHE
: * : : . :
MAB_0443|M.abscessus_ATCC_1997 IPGVD----------------------------RPPDS---DNETFVARL
TH_3646|M.thermoresistible__bu IPGRD----------------------------RPTDAAEPDSGAFVARL
MAV_0504|M.avium_104 IPGAE----------------------------RPTDSD--DPDAFVAKL
Mvan_5934|M.vanbaalenii_PYR-1 APTSS----------------------------GAAAGD----DQSLTRF
MSMEG_4156|M.smegmatis_MC2_155 LEHRDNLDRFNNPVYRLITVILMRCGLRITDALRLRGDCVTTDANGAPYL
Mflv_4644|M.gilvum_PYR-GCK IPGQG------------------------------------------SWL
. :
MAB_0443|M.abscessus_ATCC_1997 RYQQTKVEGVDPTILVEQPVVDIITEQQRWLAEHHPDIRPKYLFVGMLHQ
TH_3646|M.thermoresistible__bu RYQQTKVDGVIPTILVEQAIVDIITEQQAWLKSKYPALQPKYLFLGIKNQ
MAV_0504|M.avium_104 RYQQTKVDDVTPTILVEQAVVNVVTEQQRWLTTTYPGLTPKYLFIGVKQN
Mvan_5934|M.vanbaalenii_PYR-1 RYAQSKIDVAPDTILVDREVTEVIAEQHRWLQAQHPDGSRRYLFARRLGN
MSMEG_4156|M.smegmatis_MC2_155 RYFNHKMK-RDALVPIAPDLVDMIGYQRRLVSERWPDG-TALLFPRPTKN
Mflv_4644|M.gilvum_PYR-GCK KVPLGKLN-SERMIPVDDEVLTLVDRITTTRSSGRPMIHPRTGAPADFLF
: *:. : : : :: *
MAB_0443|M.abscessus_ATCC_1997 HQGQRPRAYSSYLNALRRLDAIHALTDSAGHKLRFTQTHRLRHTRATELL
TH_3646|M.thermoresistible__bu HRGQRPRSYTTYRAMLDKLDQCHDLTDSAGRPLRFTQTHRLRHTRATELL
MAV_0504|M.avium_104 LAGRRPRPYASYNLSLAKLDNIHGLTDAAGNPLRFTQTHRLRHTRATELL
Mvan_5934|M.vanbaalenii_PYR-1 RRGDKPYPAGTYNWVLRTLSDLVQITDAKGQVVRLSHTHRFRHTRLTRLA
MSMEG_4156|M.smegmatis_MC2_155 IDGQIPLAIPTYREALHRWLAVCDIRDEHSDPVHLTP-HQWRHTLGTRLI
Mflv_4644|M.gilvum_PYR-GCK THHGKRLSQNAVREELN--------RAAQAASLGHITPHQLRHTYATALI
. : * . : *: *** * *
MAB_0443|M.abscessus_ATCC_1997 NDGVPFHVVQRYMGHKSPEMTARYAATLAATAEAEFLKHKKIGAHGADIA
TH_3646|M.thermoresistible__bu NDGVPFHVVQRYLGHKSPEMTARYAATLAATAEAEFLKHKKIGAHGADID
MAV_0504|M.avium_104 NDGVPIHVVQRYLGHKSPEMTMRYASTLAATAEAEFLKHKKIGAHGVEIA
Mvan_5934|M.vanbaalenii_PYR-1 ELGLPIHVLQRYAGHATPTMTMHYIAARDEHAEQAFLATAKLRSDGTRIQ
MSMEG_4156|M.smegmatis_MC2_155 NRDVPQEVVRRILDHDSPQMTAHYARLHDTTVRRAWEAARKVDSHGREVN
Mflv_4644|M.gilvum_PYR-GCK NAGVSLQALMALLGHVSTQMSLRYAHLFDHTVRTEYERALDLAKSHIGAL
: .:. ..: .* :. *: :* .. : .:
MAB_0443|M.abscessus_ATCC_1997 ISPNDIYDMTQLGKR-------TDRILPNGVCLLPPLK-SCDKGNACLSC
TH_3646|M.thermoresistible__bu ITPHDIYEMTQLAKR-------TDRVLPNGVCLLPPLK-SCDKGNACLSC
MAV_0504|M.avium_104 ISPSDILDMTQLSAR-------TDRVLPNGVCLLPPLK-TCDKGNACLSC
Mvan_5934|M.vanbaalenii_PYR-1 LS-SDDHDSLHLFRR-------ADRFLPNGWCMLPPLQ-SCDKGNACLTC
MSMEG_4156|M.smegmatis_MC2_155 LDPDGPLAEAAWAKQRLGR---ATQALPNGYCGLPVQQ-SCPHANACLTC
Mflv_4644|M.gilvum_PYR-GCK PKTAVGLPITDITGTGWKDTPAIKSRLAGGYCLRAPAQGSCPYANICEHC
*..* * . : :* .* * *
MAB_0443|M.abscessus_ATCC_1997 GHFATDRTHLEELVAQRAATHTLLEQRRAQVLARTGRELTDENVWVAERL
TH_3646|M.thermoresistible__bu GHFATDATHLAELRQQLASTQTLIAQRQDQYRQRSGRELGEDNIWIIERR
MAV_0504|M.avium_104 GHFATDITHAAELRDQHTKTAALIELRRQQYRQRTGRELTDDNVWIRERL
Mvan_5934|M.vanbaalenii_PYR-1 SVFVTDHTHRDVLERQLRETSDLIDWTTTEFENRHSRPMPEDNVWLIRRH
MSMEG_4156|M.smegmatis_MC2_155 PMFLTTQEFLPQHRTQREQTLHLITAAEARGHQR----LAEMNRQVLG--
Mflv_4644|M.gilvum_PYR-GCK PSFHTDSTHLAVLAAQRIDAQDLAADAEK-----------------RGWI
* * . * : *
MAB_0443|M.abscessus_ATCC_1997 REITSLNAIIGRLSSKELGEAQAVGGPATAERIPLLPIVTRGSHQSALDK
TH_3646|M.thermoresistible__bu REIASLQAIIDRLTTEEAGSVAGAG---AAKRLPLLQIQTRGAHQSALDK
MAV_0504|M.avium_104 RELNSLQAILERLAADAGTPESAISGAGTFRRLPLHVIATRGAHDSALRK
Mvan_5934|M.vanbaalenii_PYR-1 AEHHALTKLLDALNHAPDSATHGAT--------TKCHRPSAGPVPVPLDL
MSMEG_4156|M.smegmatis_MC2_155 ----NLDAIITSLDTPTDPKAAEHAS------------------------
Mflv_4644|M.gilvum_PYR-GCK DEAERHHNLVSRLDALITGSASA---------------------------
:: *
MAB_0443|M.abscessus_ATCC_1997 ARRPDHPTG
TH_3646|M.thermoresistible__bu ARRRRTGEP
MAV_0504|M.avium_104 ADPRADA--
Mvan_5934|M.vanbaalenii_PYR-1 TRHRRRTR-
MSMEG_4156|M.smegmatis_MC2_155 ---------
Mflv_4644|M.gilvum_PYR-GCK ---------