For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRREPTKPGLPAERTLLSWERSSFGFLVGGALVLLRQHGPLGQWRAVLAITAALLALLVLALGYRRSRQI TDSPVIAGRIITSAPHTEVLVIGAATLGFATVVVAALLFAL*
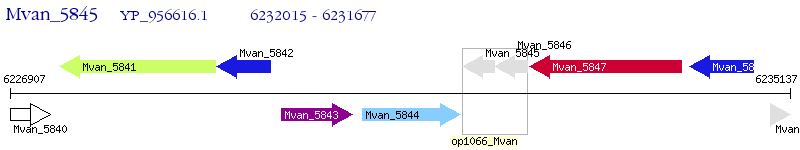
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5845 | - | - | 100% (112) | hypothetical protein Mvan_5845 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2296 | - | 3e-06 | 37.50% (104) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_0979 | - | 2e-38 | 71.43% (112) | hypothetical protein Mflv_0979 |
| M. tuberculosis H37Rv | Rv2273 | - | 3e-06 | 37.50% (104) | transmembrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3369 | - | 1e-05 | 33.66% (101) | transmembrane protein |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1230 | - | 8e-07 | 39.08% (87) | hypothetical protein MSMEG_1230 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1270 | - | 4e-06 | 33.01% (103) | transmembrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3369|M.marinum_M MSQSSETAQAADRGLQAERTTLAWTRTSFALLVNGVLLTTKDVQAHAGLA
MUL_1270|M.ulcerans_Agy99 MSQSSETAQAADRGLQAERTTLAWTRTSFALLVNGVLLTTKDVQAHAGLA
Mb2296|M.bovis_AF2122/97 MNRHS--TAASDRGLQAERTTLAWTRTAFALLVNGVLLTLKDTQGADGPA
Rv2273|M.tuberculosis_H37Rv MNRHS--TAASDRGLQAERTTLAWTRTAFALLVNGVLLTLKDTQGADGPA
MSMEG_1230|M.smegmatis_MC2_155 MTRRA---LAEDRGLQAERTALAWTRTALAIAASGVLVLLRDSHIQDDPG
Mvan_5845|M.vanbaalenii_PYR-1 MPRRE----PTKPGLPAERTLLSWERSSFGFLVGGALVLLR-QHGPLGQW
Mflv_0979|M.gilvum_PYR-GCK MPRPL----PDKPGLPAERTLLSWERSSFGFLVGGALVLLR-QHGPLGPG
* : . . ** **** *:* *:::.: ..*.*: : : .
MMAR_3369|M.marinum_M GLVPAGLAALAASCGYIIAYQRQRTLRRRPRPQR----ITARRQVYIVGL
MUL_1270|M.ulcerans_Agy99 GLVPAGLAALAASCGYTIAYQRQRTLRRRPRPQR----ITARRQVYIVGL
Mb2296|M.bovis_AF2122/97 GLIPAGLAGAAASCCYVIALQRQRALSHRPLPAR----ITPRGQVHILAT
Rv2273|M.tuberculosis_H37Rv GLIPAGLAGAAASCCYVIALQRQRALSHRPLPAR----ITPRGQVHILAT
MSMEG_1230|M.smegmatis_MC2_155 RLVVVAAVAVVALGVFALGAHRRRRLLAHPRGN------AGRRYIRAAGV
Mvan_5845|M.vanbaalenii_PYR-1 RAVLAITAALLALLVLALGYRRSRQITDSPVIAGRIITSAPHTEVLVIGA
Mflv_0979|M.gilvum_PYR-GCK RILLAVTATLLALLVLALGYRRSHAIRDSPAVT-----AEPRTEVLLMGG
: . . * :. :* : : * : : .
MMAR_3369|M.marinum_M AVLVLMVVTAFAQLL-----------------------------------
MUL_1270|M.ulcerans_Agy99 AVLVLMVVTAFAQLLSAAPGHRRPQLTRSECCSRKSAASRRTSVRSPSIC
Mb2296|M.bovis_AF2122/97 AVLVLMVVTAFAQLL-----------------------------------
Rv2273|M.tuberculosis_H37Rv AVLVLMVVTAFAQLL-----------------------------------
MSMEG_1230|M.smegmatis_MC2_155 TILAEGLLVMIYLMLA----------------------------------
Mvan_5845|M.vanbaalenii_PYR-1 ATLGFATVVVAALLFAL---------------------------------
Mflv_0979|M.gilvum_PYR-GCK ATVVFAVTVVGVLLFAVS--------------------------------
: : . ::
MMAR_3369|M.marinum_M --------------------------------------------------
MUL_1270|M.ulcerans_Agy99 ATVAARFAESVSATRCTIHDASEAVTTPIRLMPPIISNTAITRPVTVIGY
Mb2296|M.bovis_AF2122/97 --------------------------------------------------
Rv2273|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_1230|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5845|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0979|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_3369|M.marinum_M ---
MUL_1270|M.ulcerans_Agy99 KSP
Mb2296|M.bovis_AF2122/97 ---
Rv2273|M.tuberculosis_H37Rv ---
MSMEG_1230|M.smegmatis_MC2_155 ---
Mvan_5845|M.vanbaalenii_PYR-1 ---
Mflv_0979|M.gilvum_PYR-GCK ---