For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAGTNVAVVLVILDGVGARRVRADTMPTLHTLATSGAWRPDGSEAVLCSATYPNILTLVTGSTPAQHRVF ANPLFGHEFTAGAHTQTVFERASTCMSEFVVGDQYLIDVAKARTAGRHWPPEGVLPASAGVDEFGYATDA QVLPHAVAAAERAPDLLVVHFNGPDTASHLHGPDSPEAIESYRTVDAALAGFIDSLRPRWDELLVLIASD HDQEPIDDDRRIDLAGLAAARGVEAGVFHEGTAAVVVGRDACDDRWLSGVPGVEGSWLCEPGVRVIASAP HHWFAAPGYPSRGGGHGGPRTRDQVAVATGGHPMVAQIAQVWRHRRPAAQDWAGLVLAALKLATVAPR*
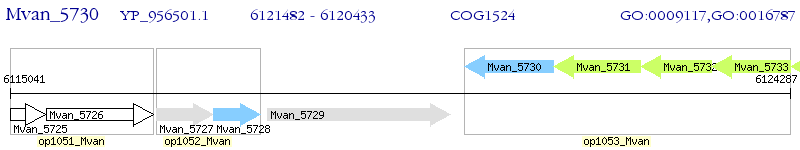
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5730 | - | - | 100% (349) | type I phosphodiesterase/nucleotide pyrophosphatase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1855 | - | 7e-05 | 33.33% (72) | hypothetical protein MMAR_1855 |
| M. avium 104 | MAV_3712 | - | 1e-06 | 26.72% (247) | type I phosphodiesterase / nucleotide pyrophosphatase |
| M. smegmatis MC2 155 | MSMEG_6672 | - | 1e-147 | 74.03% (335) | type I phosphodiesterase / nucleotide pyrophosphatase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5730|M.vanbaalenii_PYR-1 ---------------------------------MSAGTNVAVVLVILDGV
MSMEG_6672|M.smegmatis_MC2_155 ------------------------------------MTDPMVVLVIIDGA
MMAR_1855|M.marinum_M MPGSICDVLPAAGALLGVPGTEDRLG---LIDRVG--HLDRVLVVLVDGL
MAV_3712|M.avium_104 MPGSLRDVLPAAAALLGVADAGHEPGGSAVTDWVGAERVDRVLVLLVDGL
*:::::**
Mvan_5730|M.vanbaalenii_PYR-1 GARRVR--ADTMPTLHTLATSGAWRPDGSEAVLCSATYPNILTLVTGSTP
MSMEG_6672|M.smegmatis_MC2_155 GARQVR--PDTMPTLHSLATSGAWNPEGSEAVLCSATYPNILTLVTGSSP
MMAR_1855|M.marinum_M GWHLLPELAADGALLASVLAGDAGRVSQLSCTFPSTTPTSLVSLGTGAMP
MAV_3712|M.avium_104 GWQLLAQLAGDAPLLAAVRAGEVGRLTRLDCTFPSTTPTSLVSLATGAAP
* : : . . * :: :. . . ...: *:* ..:::* **: *
Mvan_5730|M.vanbaalenii_PYR-1 AQHRVFANPLFGHEFTAGAHTQTVFERASTCMS-EFVVGDQYLIDVAKAR
MSMEG_6672|M.smegmatis_MC2_155 TQHRVFANPLFGHELAGTSSVTTVFERAGDRAS-EFVVGDQYLIDVGRAR
MMAR_1855|M.marinum_M GEHGILG---FTVNVPGTDRVLNHIRWRDDPPQGQWQPLPTWFERLTWAG
MAV_3712|M.avium_104 GQHGILG---FTLRVPGTDRVLNHVRWRDDPAPDAWQPLPTWFERLAAAG
:* ::. * .... . . .. . : :: : *
Mvan_5730|M.vanbaalenii_PYR-1 TAGRHWPPEGVLPASAGVDEFG------YATDAQVLPHAVAAAERAPDLL
MSMEG_6672|M.smegmatis_MC2_155 TAGRHWPPEGVLPDDAFRDEFG------YATDCAVLPHALAAVGRRPDLL
MMAR_1855|M.marinum_M ISTRAVLPEWFIGSGLTAAAYRGAQFHPTRASDDYAQLVIDEVRGAPGLV
MAV_3712|M.avium_104 VRARAVLPSSFLGSGLTEAAYRGARLVPAEPADDYPRLVGDELRAGPGLV
* *. .: . : . . *.*:
Mvan_5730|M.vanbaalenii_PYR-1 VVHFNGPDTASHLHGPDSPEAIESYRTVDAALAGFIDSLRPRWDELLVLI
MSMEG_6672|M.smegmatis_MC2_155 VVHLNGPDTASHLHGPDSDAAVESYRTSDATLASLVDALRPHWDNLLLLV
MMAR_1855|M.marinum_M YAYTSDLDTAAHLFGIGSSQWHEAGRSVDSLLTRLVEALPP---DAALLV
MAV_3712|M.avium_104 YGYTAELDTAAHLFGIGSPQWHTAAAAVDALLSGLLEALPP---NAALLV
: ***:**.* .* : : *: *: ::::* * : :*:
Mvan_5730|M.vanbaalenii_PYR-1 ASDHDQEPIDDDRRIDLAG-LAAARGVEAGVFHEGTAAVVVGRDACDDRW
MSMEG_6672|M.smegmatis_MC2_155 TSDHDQETIHDDRRIDLAG-LASARGVDATVFHEGTAAVICGPGAADARW
MMAR_1855|M.marinum_M TADHGGLNVAADARVDLDTDPRLSAGIRVVAGEPRVRYLHTVPGATTDVL
MAV_3712|M.avium_104 TADHGGLNVPPEARVDLDSDPRLAAGIRVVAGEPRVRYLHTEPGAAPDVQ
::**. : : *:** : *: . . . . : .*
Mvan_5730|M.vanbaalenii_PYR-1 LSGVPGVEGSWLCEPGVRVIASAPHHWFAAPGYPSRGGGHGGPRTRDQVA
MSMEG_6672|M.smegmatis_MC2_155 LSDVAGIERSWLVEPGLRVAASEPHHWFATADGPTRGGAHGGPRTRGQVA
MMAR_1855|M.marinum_M DTWRELLGTRADVRTRDDAIATGIFGSVAPEHRLRIGDVVVTCADDAAVL
MAV_3712|M.avium_104 AAWSELLDGRAAVYSRAQAVATGMFGPVADRHLPRLGDVVVVCSADTAVL
: : . . *: . .* *. *
Mvan_5730|M.vanbaalenii_PYR-1 VATGGHPMVAQIAQVWRHRRPAAQDWAGLVLAALKLATVAPR
MSMEG_6672|M.smegmatis_MC2_155 VAAGGHPMVGRIAGAWRHRQPRAEDWAGLALSVFG-------
MMAR_1855|M.marinum_M ATAHEPPETARLIGFHGAATAAEMAIPLISF-----------
MAV_3712|M.avium_104 ASAHEPPETARLVGFHGAATPAETAIPLILFRR---------
.:: * ..:: . . : :