For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDPMVVLVIIDGAGARQVRPDTMPTLHSLATSGAWNPEGSEAVLCSATYPNILTLVTGSSPTQHRVFAN PLFGHELAGTSSVTTVFERAGDRASEFVVGDQYLIDVGRARTAGRHWPPEGVLPDDAFRDEFGYATDCAV LPHALAAVGRRPDLLVVHLNGPDTASHLHGPDSDAAVESYRTSDATLASLVDALRPHWDNLLLLVTSDHD QETIHDDRRIDLAGLASARGVDATVFHEGTAAVICGPGAADARWLSDVAGIERSWLVEPGLRVAASEPHH WFATADGPTRGGAHGGPRTRGQVAVAAGGHPMVGRIAGAWRHRQPRAEDWAGLALSVFG
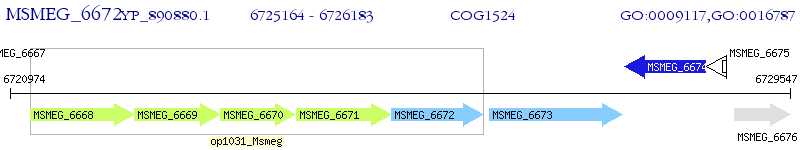
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6672 | - | - | 100% (339) | type I phosphodiesterase / nucleotide pyrophosphatase superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3271 | - | 6e-05 | 32.26% (124) | type I phosphodiesterase/nucleotide pyrophosphatase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1855 | - | 8e-06 | 39.44% (71) | hypothetical protein MMAR_1855 |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_1346 | - | 9e-06 | 34.68% (124) | phosphodiesterase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5730 | - | 1e-147 | 74.03% (335) | type I phosphodiesterase/nucleotide pyrophosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3271|M.gilvum_PYR-GCK MKLPRPDPAQPHLADVAPSVLAAMGVPGFEPRIDLPFDIG---GACVLLV
TH_1346|M.thermoresistible__bu MDLPRPDPGLAHLADVVPAVLAAMGVAGFDSRIPLRHGLA---GACVLLI
MMAR_1855|M.marinum_M --MPG------SICDVLPAAGALLGVPGTEDRLGLIDRVGHLDRVLVVLV
MSMEG_6672|M.smegmatis_MC2_155 ---------------MTDPMVVLVIIDGAGARQVRPDTMP----------
Mvan_5730|M.vanbaalenii_PYR-1 ------------MSAGTNVAVVLVILDGVGARRVRADTMP----------
. : : * * :
Mflv_3271|M.gilvum_PYR-GCK DGLGAELLDRYADD----APVLAGMRG--ATLNVGFPSTTAAGLAAVGTG
TH_1346|M.thermoresistible__bu DGLGAELLDTHADD----APVLAGLRG--ETLQVGFPSTTAAGLAALGTG
MMAR_1855|M.marinum_M DGLGWHLLPELAADGALLASVLAGDAGRVSQLSCTFPSTTPTSLVSLGTG
MSMEG_6672|M.smegmatis_MC2_155 ------TLHSLATS---------GAWNP-EGSEAVLCSATYPNILTLVTG
Mvan_5730|M.vanbaalenii_PYR-1 ------TLHTLATS---------GAWRP-DGSEAVLCSATYPNILTLVTG
* * . * . : *:* ..: :: **
Mflv_3271|M.gilvum_PYR-GCK CRSGEHGMVG---MTFRLPDVG-VVNALRWRPHPWGDDLRDRVTPETVQP
TH_1346|M.thermoresistible__bu CHSGEHGFVG---YSFRVPAAG-VINALRWRPHPWGEDLRAELPPEQVQP
MMAR_1855|M.marinum_M AMPGEHGILG---FTVNVPGTDRVLNHIRWRDDP---------PQGQWQP
MSMEG_6672|M.smegmatis_MC2_155 SSPTQHRVFANPLFGHELAGTSSVTTVFER---------------AGDRA
Mvan_5730|M.vanbaalenii_PYR-1 STPAQHRVFANPLFGHEFTAGAHTQTVFER---------------ASTCM
. . :* ... ... . . :.
Mflv_3271|M.gilvum_PYR-GCK RPTLFERAAATGVAVSVISGAQFAGSGLTRAVLRGGAYVGVHTMGDLAAT
TH_1346|M.thermoresistible__bu LPTTFERAAAAGVAVSVISGAQFTGSGLTRAVLRGGRYVGVHALGDLAAT
MMAR_1855|M.marinum_M LPTWFERLTWAGISTRAVLPEWFIGSGLTAAAYRGAQFHPTRASDDYAQL
MSMEG_6672|M.smegmatis_MC2_155 SEFVVGDQYLIDVGRARTAGRHWPPEGVLPDDAFRDEFGYATDCAVLPHA
Mvan_5730|M.vanbaalenii_PYR-1 SEFVVGDQYLIDVAKARTAGRHWPPEGVLPASAGVDEFGYATDAQVLPHA
. .:. : .*: : . .
Mflv_3271|M.gilvum_PYR-GCK IRSVLADR-GFCYGYHADLDLVGHLHGPGSEAWRLQLRQVDRLVESIVEG
TH_1346|M.thermoresistible__bu VRQTIADG-GFCYGYHSDVDLLGHLHGPGSPAWRMQLRQVDRLVESVLDD
MMAR_1855|M.marinum_M VIDEVRGAPGLVYAYTSDLDTAAHLFGIGSSQWHEAGRSVDSLLTRLVEA
MSMEG_6672|M.smegmatis_MC2_155 LAAVGRRP-DLLVVHLNGPDTASHLHGPDSDAAVESYRTSDATLASLVDA
Mvan_5730|M.vanbaalenii_PYR-1 VAAAERAP-DLLVVHFNGPDTASHLHGPDSPEAIESYRTVDAALAGFIDS
: .: : . * .**.* .* * * : .::
Mflv_3271|M.gilvum_PYR-GCK LPR---GGLLAVVADHGMIAL-DDNAVDLDVSAALLAGTEAIGGEVRARH
TH_1346|M.thermoresistible__bu LPS---GGLLAVVADHGMVGVTDDHVIDLDAHVTLWDGVDAVAGEPRARH
MMAR_1855|M.marinum_M LPP---DAALLVTADHGGLNVAADARVDLDTDPRLSAGIRVVAGEPRVRY
MSMEG_6672|M.smegmatis_MC2_155 LRPHWDNLLLLVTSDHDQETIHDDRRIDLAG-LASARGVDATVFHEGTAA
Mvan_5730|M.vanbaalenii_PYR-1 LRPRWDELLVLIASDHDQEPIDDDRRIDLAG-LAAARGVEAGVFHEGTAA
* : :.:**. : * :** * . . .
Mflv_3271|M.gilvum_PYR-GCK VYTHPGAVDDVLAAWQSTLGDRAWVVSRDEAIASGWFGERVADDVRSRIG
TH_1346|M.thermoresistible__bu VYAADGAADDVLATWRDTLGEQAWVLSREEAVTAGWFGHRVTDEVRYRIG
MMAR_1855|M.marinum_M LHTVPGATTDVLDTWRELLGTRADVRTRDDAIATGIFGS-VAPEHRLRIG
MSMEG_6672|M.smegmatis_MC2_155 VICGPGAADARWLSDVAGIERSWLVEPGLRVAASEPHHWFATADGPTRGG
Mvan_5730|M.vanbaalenii_PYR-1 VVVGRDACDDRWLSGVPGVEGSWLCEPGVRVIASAPHHWFAAPGYPSRGG
: .* : : . . :: . .: * *
Mflv_3271|M.gilvum_PYR-GCK DVVAAARGCAGMLRRATEPVESS-LVGQHGSLTAAEQTVPLLLAHR----
TH_1346|M.thermoresistible__bu DVVAAARGTAALVRRSAEPLESA-LIGHHGSLTTAEQRVPLLLAYG----
MMAR_1855|M.marinum_M DVVVTCADDAAVLATAHEPPETARLIGFHGAATAAEMAIPLISF------
MSMEG_6672|M.smegmatis_MC2_155 AHGGPRTRGQVAVAAGGHPMVGRIAGAWRHRQPRAEDWAGLALSVFG---
Mvan_5730|M.vanbaalenii_PYR-1 GHGGPRTRDQVAVATGGHPMVAQIAQVWRHRRPAAQDWAGLVLAALKLAT
. : . .* : . *: *
Mflv_3271|M.gilvum_PYR-GCK ----
TH_1346|M.thermoresistible__bu ----
MMAR_1855|M.marinum_M ----
MSMEG_6672|M.smegmatis_MC2_155 ----
Mvan_5730|M.vanbaalenii_PYR-1 VAPR