For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAFDITPTAAQHDLARRTHEFAEEVIRPVAADYDQRQEFPWPVLEEAAARGFYSPLFYRDLIGDPTGLSL PMFMEELFWGCAGIGLAVVMPALALSAIGQAASPEQMLQWAPECFGTPGDLKLAALAISEPEGGSDVRNL RTTARRAGPGADADWIIDGHKMWIGNGGIADVHVVNAVVDQELGHKGQALFIVPGGTPGLDMVRKLDKLG CRASHTAELKLTGVRVPADHLLGGQEKLDHKLARAREAIEGARHSGSATLGTFEQTRPMVAAQALGIARA ALEYVTEYANRREAFGAPIIDNQGISFPLADLATGLDAARLLTWRASWMAATGVPFERGEGSMSKLAASE LAVRATERAVQTMGGWGYVKDHPVEKWYRDAKLYTIFEGTSEIQRMVISNALGAADGKPPLHVDLEPTGG PLNRMFGRGTPARTQVAASALAMKDRVPAPVMQMAMKVLRPPRK
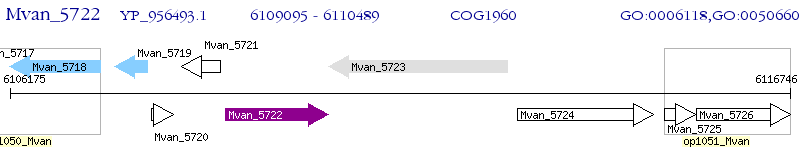
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5722 | - | - | 100% (464) | acyl-CoA dehydrogenase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2567 | - | e-107 | 49.49% (390) | acyl-CoA dehydrogenase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1719 | - | 1e-49 | 34.23% (409) | acyl-CoA dehydrogenase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3302c | fadE25 | 1e-48 | 33.91% (404) | acyl-CoA dehydrogenase FADE25 |
| M. gilvum PYR-GCK | Mflv_1095 | - | 0.0 | 92.89% (464) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3274c | fadE25 | 1e-48 | 33.91% (404) | acyl-CoA dehydrogenase FADE25 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 1e-51 | 35.32% (402) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3618c | - | 8e-52 | 35.24% (403) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_0149 | fadE25_2 | 1e-110 | 51.56% (384) | acyl-CoA dehydrogenase FadE25_2 |
| M. avium 104 | MAV_0039 | - | 0.0 | 84.63% (462) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0531 | - | 1e-110 | 52.34% (384) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2926 | - | 0.0 | 84.27% (464) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4948 | fadE25_2 | 1e-109 | 51.30% (384) | acyl-CoA dehydrogenase FadE25_2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5722|M.vanbaalenii_PYR-1 --------MAFDITPTAAQHDLARRTHEFAEEVIRPVAADYDQRQEFPWP
Mflv_1095|M.gilvum_PYR-GCK --------MTFDITPTAAQHDLARRAHEFAEDVVRPVAAEYDQRQEFPWP
MAV_0039|M.avium_104 --------MTFDLTPTAAQHDLARRTHEFAESVIRPVALEYDQRQEFPWP
TH_2926|M.thermoresistible__bu ------MAMAFDLTPTVAQHDLAARTHEFAEQVIRPVASEYDQRQEFPWP
MMAR_0149|M.marinum_M --------MTFSLELSDDVIEVRDWVHKFSAEVIRPAAAEWDEREETPWP
MUL_4948|M.ulcerans_Agy99 --------MTFSLELSDDVIEVRDWVHKFSAEVIRPAAAEWDEREETPWP
MSMEG_0531|M.smegmatis_MC2_155 --------MTFSLQLSDDVIEVRDWVHQFAAEVVRPAAAEWDEREETPWP
Mb3302c|M.bovis_AF2122/97 MVGWAGNPSFDLFKLPEEHDEMRSAIRALAEKEIAPHAAEVDEKARFPEE
Rv3274c|M.tuberculosis_H37Rv MVGWAGNPSFDLFKLPEEHDEMRSAIRALAEKEIAPHAAEVDEKARFPEE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
MAB_3618c|M.abscessus_ATCC_199 ---MAGNPSFDLFKLGEEHDELRAAIRGLAEKEIAPHAKDVDEKARFPQE
: :: : :: . : * * : *:: . *
Mvan_5722|M.vanbaalenii_PYR-1 VLEEAAARGFYSPLFYRDLIGD-PTGLSLPMFMEELFWGCAGIGLAVVMP
Mflv_1095|M.gilvum_PYR-GCK VLEEAATRGFYSPLFYRDLIGD-PTGLSLPMFMEELFWGCAGIGLAVVMP
MAV_0039|M.avium_104 VLEEAARRGFYSPLFYRDLIGD-PTGISLPMFMEELFWGCAGIGLAIVMP
TH_2926|M.thermoresistible__bu VLEEAATRGFYSPLFYRDLIGD-PTGLSLPMFMEELFWGCAGIGLAIVMP
MMAR_0149|M.marinum_M VIQEAAKVGLYSPDFFAQQSVE-PSGVGILAAFEEMFWGDAGIAISIMGT
MUL_4948|M.ulcerans_Agy99 VIQEATKVGLYSPDFFAQQSVE-PSGVGILAAFEEMFWGDAGIAISIMGT
MSMEG_0531|M.smegmatis_MC2_155 VIQEAAKVGLYSPELFAQQAAE-PTGIGMLTVFEELFWGDAGIALSILGT
Mb3302c|M.bovis_AF2122/97 ALVALNSSGFNAVHIPEEYGGQGADSVATCIVIEEVARVDASASLIPAVN
Rv3274c|M.tuberculosis_H37Rv ALVALNSSGFNAVHIPEEYGGQGADSVATCIVIEEVARVDASASLIPAVN
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
MAB_3618c|M.abscessus_ATCC_199 ALDALVASGFNAVHVPEEYDGQGADSVAACIVIEEVARVCASSSLIPAVN
.: *: : . : : . .:. :**: *. .:
Mvan_5722|M.vanbaalenii_PYR-1 ALALSAIGQAASPEQMLQWAPECFGTPGDLKLAALAISEPEGGSDVRNLR
Mflv_1095|M.gilvum_PYR-GCK ALALSAIGQAASPEQMLQWAPECFGTPGDLKLAALAISEPEGGSDVRNLR
MAV_0039|M.avium_104 ALALSAIGQAASPEQMLEWAPECFGTPGDLKLAALAISEPEGGSDVRNLR
TH_2926|M.thermoresistible__bu ALALSAIGQAATPEQMLRWAPECFGTPGDLKLAALAISEPEGGSDVRNLR
MMAR_0149|M.marinum_M GLAAAALAANGTPEQLGRWLPEMFGTPGDPKLGAFCSSEPDAGSDVGAIR
MUL_4948|M.ulcerans_Agy99 GLAAAALAANGTPEQLGRWLPEMFGTPGDPKLGAFCSSEPGAGSDVGAIR
MSMEG_0531|M.smegmatis_MC2_155 GLAAAALAGNGTPEQLGQWLPEMFGTADEPKLGAFCSSEPDAGSDVGAIR
Mb3302c|M.bovis_AF2122/97 KLGTMGLILRGSEELKKQVLPALAAEG---AMASYALSEREAGSDAASMR
Rv3274c|M.tuberculosis_H37Rv KLGTMGLILRGSEELKKQVLPALAAEG---AMASYALSEREAGSDAASMR
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEG---AMASYALSEREAGSDAASMR
MAB_3618c|M.abscessus_ATCC_199 KLGTMGLILNGSDELKKQVLPAIASGE---AMASYALSEREAGSDAASMR
*. .: .: * . * . :.: . ** .***. :*
Mvan_5722|M.vanbaalenii_PYR-1 TTARRAGPGADADWIIDGHKMWIGNGGIADVHVVNAVVDQELGHKGQALF
Mflv_1095|M.gilvum_PYR-GCK TTARRAGPEPDADWIIDGHKMWIGNGGIANVHVVNAVVDQELGHKGQALF
MAV_0039|M.avium_104 TRARRDGD----DWIIDGHKMWIGNGGIANVHVVNAVVDEELGHRGQALF
TH_2926|M.thermoresistible__bu TRAVRADSGPDADWIIDGHKMWIGNGGIANVHVVNAVVDEELGHRGQALF
MMAR_0149|M.marinum_M TRAHFDEAAG--EWVLNGTKTWATNGGIANVHIVVVSVHPELGTRGQATF
MUL_4948|M.ulcerans_Agy99 TRAHFDEAAG--EWVLNGTKTWATNGGIANVHIVVVSVHPELGTRGQATF
MSMEG_0531|M.smegmatis_MC2_155 TRARFDEATR--EWVLNGTKTWATNGGIANVHIVVASVYPELGSRGQATF
Mb3302c|M.bovis_AF2122/97 TRAKADGD----HWILNGAKCWITNGGKSTWYTVMAVTDPDRGANGISAF
Rv3274c|M.tuberculosis_H37Rv TRAKADGD----HWILNGAKCWITNGGKSTWYTVMAVTDPDRGANGISAF
MLBr_00737|M.leprae_Br4923 TRAKADGD----DWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAF
MAB_3618c|M.abscessus_ATCC_199 TRAKADGD----DWIINGSKCWITNGGKSSWYTVMAVTDPEKKANGISAF
* * .*:::* * * *** : : * . . : .* : *
Mvan_5722|M.vanbaalenii_PYR-1 IVPGGTPGLDMVRKLDKLGCRASHTAELKLTGVRVPADHLLGGQEKLDHK
Mflv_1095|M.gilvum_PYR-GCK VVPGGTPGLDMVRKLDKLGCRASHTAELKFNSVRVPADHLLGGQEKLDHK
MAV_0039|M.avium_104 VVPGGTPGLKMVRKLDKLGCRASHTAELLFEGVRVPAANLLGGQEKLEHK
TH_2926|M.thermoresistible__bu VVPGGTPGLQLVRKLDKLGCRASHTAELRFTGVRVPADHLLGGQDKLEYK
MMAR_0149|M.marinum_M VIPPGTKGLTQGQKFKKHGIRASHTAEVVLDNVRLPEDAILGGREKFEAR
MUL_4948|M.ulcerans_Agy99 VIPPGTKGLTQGQKFKKHGIRASHTAEVVLDNVRLPEDAILGGREKFETR
MSMEG_0531|M.smegmatis_MC2_155 IVPPGTHGLVQGQKFKKHGIRASHTAEVVLDNVRLPEDMILGGREKFEER
Mb3302c|M.bovis_AF2122/97 MVHKDDEGFTVGPKERKLGIKGSPTTELYFENCRIPGDRIIGEPG-----
Rv3274c|M.tuberculosis_H37Rv MVHKDDEGFTVGPKERKLGIKGSPTTELYFENCRIPGDRIIGEPG-----
MLBr_00737|M.leprae_Br4923 IVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPG-----
MAB_3618c|M.abscessus_ATCC_199 MVHKDDEGFTVGPLEHKLGIKGSPTAELYFENCRIPGDRIIGEPG-----
:: . *: * * :.* *:*: : *:* ::*
Mvan_5722|M.vanbaalenii_PYR-1 LARAREAIEGARHSGSATLGTFEQTRPMVAAQALGIARAALEYVTEYANR
Mflv_1095|M.gilvum_PYR-GCK LARAREAVEGGRHSGSATLGTFEQTRPMVAAQALGIARAALEYVTEYANR
MAV_0039|M.avium_104 LAKAREVVAGGKRSGSATLGTFEQTRPMVAAQAIGIARAALEYATSYATE
TH_2926|M.thermoresistible__bu LAKARETAADGRRSGSATLGTFEQTRPMVAAQALGIARAALEYMTDYANR
MMAR_0149|M.marinum_M IARVKSGASAR---GQAAMKTFERTRPTVGAMAVGVARAAYEYALEYACQ
MUL_4948|M.ulcerans_Agy99 IARVKSGASAR---GQAAMKTFERTRPTVGAMAVGVARAAYEYALEYACQ
MSMEG_0531|M.smegmatis_MC2_155 IARVKSGASAG---GHAAMKTFERTRPTVGAMAVGVARAAYEYALDYACQ
Mb3302c|M.bovis_AF2122/97 ------------TGFKTALATLDHTRPTIGAQAVGIAQGALDAAIAYTKD
Rv3274c|M.tuberculosis_H37Rv ------------TGFKTALATLDHTRPTIGAQAVGIAQGALDAAIAYTKD
MLBr_00737|M.leprae_Br4923 ------------TGFKTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKD
MAB_3618c|M.abscessus_ATCC_199 ------------TGFKTALQTLDHTRPTIGAQALGIAQGALDQAIAYTKD
::: *:::*** :.* *:*:*:.* : *:
Mvan_5722|M.vanbaalenii_PYR-1 REAFGAPIIDNQGISFPLADLATGLDAARLLTWRASWMAATGVPFERGEG
Mflv_1095|M.gilvum_PYR-GCK REAFGGPIIDNQGISFPLADLATGLDAARLLTWRASWMAATGVPFERGEG
MAV_0039|M.avium_104 REAFGAPIIDNQGIAFPLAELATQIDAARLLTWRASWMAANNVPFERGEG
TH_2926|M.thermoresistible__bu REAFGAPIIDNQGIAFPIAEIATRIDAARLLTWRASWMAATGVPFGRGEG
MMAR_0149|M.marinum_M REQFGRKIGEFQAVAFKLADMKARIDAARLLVWRAGWMARNNHNFDSAEG
MUL_4948|M.ulcerans_Agy99 REQFGRKIGEFQAVAFKLADMKARIDAARLLVWRAGWMARNNHNFDSAEG
MSMEG_0531|M.smegmatis_MC2_155 REQFGRKIGEFQAVAFKLADMKSRIDAARLLVWRAGWMARNNQQFDSAEG
Mb3302c|M.bovis_AF2122/97 RKQFGESISTFQAVQFMLADMAMKVEAARLMVYSAAARAERGEPDLGFIS
Rv3274c|M.tuberculosis_H37Rv RKQFGESISTFQAVQFMLADMAMKVEAARLMVYSAAARAERGEPDLGFIS
MLBr_00737|M.leprae_Br4923 RKQFGESISTFQSIQFMLADMAMKVEAARLIVYAAAARAERGEPDLGFIS
MAB_3618c|M.abscessus_ATCC_199 RKQFGKSISDFQGVQFMLADMAMKVEAARLMVYTAAARAERGEKNLGFIS
*: ** * *.: * :*:: ::****:.: *. * . .
Mvan_5722|M.vanbaalenii_PYR-1 SMSKLAASELAVRATERAVQTMGGWGYVKDHPVEKWYRDAKLYTIFEGTS
Mflv_1095|M.gilvum_PYR-GCK SMSKLAASELAVRATERAIQTMGGYGYVKDHPVEKWYRDAKLYTIFEGTS
MAV_0039|M.avium_104 SMSKMAASEVAVKATERAVQTLGGWGYITDHPVEKWYRDAKLYTIFEGTS
TH_2926|M.thermoresistible__bu SMAKMAASEIAVRATEQAIQTMGGWGYITDHPVEKWYRDAKLYTIFEGTS
MMAR_0149|M.marinum_M SMAKLVASETAVYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTS
MUL_4948|M.ulcerans_Agy99 SMAKLVASETAVYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTS
MSMEG_0531|M.smegmatis_MC2_155 SMAKLVASETAVYVTDEAIQILGGNGYTRDYPVERMHRDAKIFTIFEGTS
Mb3302c|M.bovis_AF2122/97 AASKCFASDVAMEVTTDAVQLFGGAGYTTDFPVERFMRDAKITQIYEGTN
Rv3274c|M.tuberculosis_H37Rv AASKCFASDVAMEVTTDAVQLFGGAGYTTDFPVERFMRDAKITQIYEGTN
MLBr_00737|M.leprae_Br4923 AASKCFASDIAMEVTTDAVQLFGGAGYTSDFPVERFMRDAKITQIYEGTN
MAB_3618c|M.abscessus_ATCC_199 SASKCFASDVAMEVTTDAVQLFGGAGYTTDFPVERMMRDAKITQIYEGTN
: :* **: *: .* *:* :** ** *.***: ****: *:***.
Mvan_5722|M.vanbaalenii_PYR-1 EIQRMVISNALGAADGKPPLHVDLEPTGGPLNRMFGRGTPARTQVAASAL
Mflv_1095|M.gilvum_PYR-GCK EIQRIVISNALGAAEGRPPLHVDLDPTGGPFNRLLGRGTPARTRAGARAL
MAV_0039|M.avium_104 EIQRMVVSNALGAAVGTPPLHVVLEPSGGALNRVFGRGTPLRSRAAEAAL
TH_2926|M.thermoresistible__bu EIQRVVISNALGAAAGKPPLHFELDPSGGPLNRYLGRGTPLRGRASDAML
MMAR_0149|M.marinum_M EIQRLVISRAL-------------------------TGLPIR--------
MUL_4948|M.ulcerans_Agy99 EIQRLVISRAL-------------------------TGLPIR--------
MSMEG_0531|M.smegmatis_MC2_155 EIQRLVISRAL-------------------------TGLAIR--------
Mb3302c|M.bovis_AF2122/97 QIQRVVMSRALLR-------------------------------------
Rv3274c|M.tuberculosis_H37Rv QIQRVVMSRALLR-------------------------------------
MLBr_00737|M.leprae_Br4923 QIQRVVMSRALLR-------------------------------------
MAB_3618c|M.abscessus_ATCC_199 QIQRVVMSRALLTS------------------------------------
:***:*:*.**
Mvan_5722|M.vanbaalenii_PYR-1 AMKDRVPAPVMQMAMKVLRPPRK
Mflv_1095|M.gilvum_PYR-GCK TLKDRVPAPVMQMAMKVLKPPRK
MAV_0039|M.avium_104 SMQDRIPEPVMRLAMKVLRPPGR
TH_2926|M.thermoresistible__bu AARDRIPEPVMRLAMKVLRPPRR
MMAR_0149|M.marinum_M -----------------------
MUL_4948|M.ulcerans_Agy99 -----------------------
MSMEG_0531|M.smegmatis_MC2_155 -----------------------
Mb3302c|M.bovis_AF2122/97 -----------------------
Rv3274c|M.tuberculosis_H37Rv -----------------------
MLBr_00737|M.leprae_Br4923 -----------------------
MAB_3618c|M.abscessus_ATCC_199 -----------------------