For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SQATIEDAPPRTGVRVRVQKLGTALSNMVMPNIGAFIAWGLITALFIEQGWLQGIFAELKNPDGWVAKIG GWGAYDGAGIVGPMITYLLPVLIGYTGGRMVHGNRGAVVGAIATMGVVTGADVPMFLGAMIMGPLGGWCM KKLDALWEGKIRPGFEMLVDNFSAGILGLILAVFGFFGIGPIVSSFTRAAGNAVDFLVENDLLPLTSILI EPAKVLFLNNAINHGVLTPLGTTQALETGKSILFLLEANPGPGLGLLLAYMAFGRGLARASAPGAAIIQF FGGIHEIYFPYVLMKPKLIVATILGGMTGVFINVLFGSGLRAPAAPGSIIAVYAQTASGSFLGVTLSVFG SAAVSFAVAALLLKTDRATDESDLAAATAEMESLKGKKSSVAGALVATSTGPIRSIVFACDAGMGSSAMG ASVLRRKIQQAGFGDVKVTNSAISNLSDTYDLVISHRDLTARARQRTGSAVHVSVDDFMGSPRYDEIVER LKQTNTGGGDGAPAAVEEPAEAPADDVLPLSSIVLDGAATTAADAITEAGRLLVTAGAVEPAYIDAMHQR ESSISTYMGNGLAIPHGTNEAKDTIRRTGLSFVRYAEPIDWNGKPAEFVVGIAGAGKDHMALLTKIAQVF LKADDVARLREATTTEEVKAILMAADK*
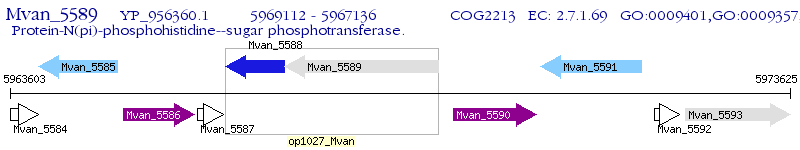
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5589 | - | - | 100% (658) | PTS system, mannitol-specific IIC subunit |
| M. vanbaalenii PYR-1 | Mvan_0094 | - | 5e-07 | 29.58% (142) | PTS system, fructose subfamily, IIC subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1219 | - | 0.0 | 84.85% (660) | PTS system, mannitol-specific IIC subunit |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0085 | - | 3e-08 | 33.08% (130) | PTS system, Fru family protein, IIABC components |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5589|M.vanbaalenii_PYR-1 MSQATIEDAPPRTGVRVR------VQKLGTALSNMVMPNIGAFIAWGLIT
Mflv_1219|M.gilvum_PYR-GCK MSQTTVDDAPARTGVRVR------VQKLGTALSNMVMPNIAAFIAWGLIT
MSMEG_0085|M.smegmatis_MC2_155 MTQPIITDDLVLLDVDAGSDKEAVIARLAGALGGAGRASDTAGLVGAAMA
*:*. : * .* . : :*. **.. .. * :. . ::
Mvan_5589|M.vanbaalenii_PYR-1 ALFIEQGWLQG--IFAELKNPDGWVAKIGGWGAYDGAGIVGPMITYLLPV
Mflv_1219|M.gilvum_PYR-GCK ALFIEQGWLQG--IFAGLKDPDGWVAKIGGWGAYDGAGIVGPMITFLLPI
MSMEG_0085|M.smegmatis_MC2_155 REAQSATGLPGGIAIPHCRSPYVDTATIGFARLDPKVDFGAPDGPADLAF
. * * :. :.* .*.** ..: .* . *..
Mvan_5589|M.vanbaalenii_PYR-1 LIGYTGG------RMVHGNRGAVVGAIATMGVVTGADVPMFLGAMIMGPL
Mflv_1219|M.gilvum_PYR-GCK LIGYTGG------RMIHGNRGAVVGAIATVGVVTGADVPMFLGAMIMGPL
MSMEG_0085|M.smegmatis_MC2_155 LIAAPESGGSEHMKLLSSLARALVRKDFVESLRSASSTQEVVELVNSAVK
**. . . ::: . *:* . .: :.:.. .: : .
Mvan_5589|M.vanbaalenii_PYR-1 GGWCMKKLDALWEGKIRPGFEMLVDNFSAGILGLILAVFGFFGIGPIVSS
Mflv_1219|M.gilvum_PYR-GCK GGWCMKKIDAIWDGKIRPGFEMLVDNFSAGILGMILAVFGFFGIGPIVSA
MSMEG_0085|M.smegmatis_MC2_155 PAAAPPASAPKPAAEPKPAADKSVSIVAVTSCPTGIAHTYMAADALKAAA
. . . .: :*. : *. .:. :* : . . .::
Mvan_5589|M.vanbaalenii_PYR-1 FTRAAGNAVDFLVENDLLPLTSILIEPAKVLFLNNAINHGVLTPLGTTQA
Mflv_1219|M.gilvum_PYR-GCK FTRGAGNAVDFLVDNDLLPLTSILIEPAKVLFLNNAINHGVLTPLGTTQA
MSMEG_0085|M.smegmatis_MC2_155 EKAGVACTVETQGSAGSTPLSAETIAAADAVIFATDVGVKDRQRFAGKPV
. ... :*: . . **:: * .*..::: . :. :. . .
Mvan_5589|M.vanbaalenii_PYR-1 LETGKSILFLLEANPGPGLGLLLAYMAFGRGLARASAPGAAIIQFFGGIH
Mflv_1219|M.gilvum_PYR-GCK LETGKSILFLLETNPGPGLGILLAFTVFGKGVARASAPGAAIIQFFGGIH
MSMEG_0085|M.smegmatis_MC2_155 IASG-VKRAINEPDKMVAEAISAASDPGAPWVAGQAAESASAPAGDVGWG
: :* : *.: . .: * . :* :* .*: *
Mvan_5589|M.vanbaalenii_PYR-1 EIYFPYVLMKPKLIVATILGGMTGVFINVLFGSGLRAPAAPGSIIAVYAQ
Mflv_1219|M.gilvum_PYR-GCK EIYFPYVLMKPKLIIATILGGMTGVFINVLFGSGLRAPAAPGSIIAIYAQ
MSMEG_0085|M.smegmatis_MC2_155 TRIRQILLTGVSYMIPFVAAGGLLIALGFLLAGYEIGNTPDGETQSLGNI
:* . ::. : .* : :..*:.. . :. *. ::
Mvan_5589|M.vanbaalenii_PYR-1 TASGSFLGVTLSVFGSAAVSFAVAALLLKTDRATDESDLAAATAEMESLK
Mflv_1219|M.gilvum_PYR-GCK TASGSFLGVTLSVLGAATVSFVVAAFLLKTDKADDEQDLAAATASMESMK
MSMEG_0085|M.smegmatis_MC2_155 IAT----HNSLTNLPSGGVAQYLGAVLFTLGGLAFSFLVPALAGYIAFAI
*: :*: : :. *: :.*.*:. . . :.* :. :
Mvan_5589|M.vanbaalenii_PYR-1 GKKSSVAGALVATSTG----PIRSIVFACDAGMGSSAMGASVLRRKIQQA
Mflv_1219|M.gilvum_PYR-GCK GKKSSVSSQLVGASSGGPRGPIKSIVFACDAGMGSSAMGASVLRRKIQQA
MSMEG_0085|M.smegmatis_MC2_155 ADRPGIAPGFTAGAVAVFVGGGFIGGIVGGLIAGFAALWISRIGVPQWAR
..:..:: :.. : . :. . * :*: * :
Mvan_5589|M.vanbaalenii_PYR-1 GFGDVKVTNSAISNLSDTYDLVISHRDLTARARQRTGSAVHVSVDDFMGS
Mflv_1219|M.gilvum_PYR-GCK GFGDVKVTNSAISNLTDSYDLVVSHRDLTARAGQRTASAIHVSVDDFMSS
MSMEG_0085|M.smegmatis_MC2_155 GLMPVVVIPLFASMVVGLLMFLLLGRPLAALTSGLTNWLGGLSGGSAIAL
*: * * * : . ::: * *:* : * :* .. :.
Mvan_5589|M.vanbaalenii_PYR-1 PRYDEIVERLKQTNTGGGDGAPAAVEEPAEAPADDVLPLSSIVLDGAATT
Mflv_1219|M.gilvum_PYR-GCK PRYDEIVEQLKQGNGGGSEAEPMVEDGSAGSSDGDVLALSSIVLNGSASS
MSMEG_0085|M.smegmatis_MC2_155 GVILGLMMCFDLGGPVNKAAYAFATASLVNPTQANLQIMAAVMAAGMVPP
:: :. . . . . . . .. :: ::::: * ...
Mvan_5589|M.vanbaalenii_PYR-1 AADAITEAGRLLVTAGAVEPAYIDAMHQRESSISTYMGNGLAIPHGTNEA
Mflv_1219|M.gilvum_PYR-GCK AADAISEAGRLLVDAGAVEPSYVDAMHERESSVSTYMGNGLAIPHGTNEA
MSMEG_0085|M.smegmatis_MC2_155 LAMALATVIRPRLFSEPERENGRAAWLLGASFITEGAIPFAAADPLRVIP
* *:: . * : : . . * * :: * .
Mvan_5589|M.vanbaalenii_PYR-1 KDTIRRTGLSFVRYAEPIDWNGKPAEFVVGIAGAGKDHMALLTKIAQVFL
Mflv_1219|M.gilvum_PYR-GCK KGAIRRTGLSLVRYPEPIDWNGKPAEFVVGIAGAGKDHMALLSKIAQVFS
MSMEG_0085|M.smegmatis_MC2_155 SMMVGGAVTGATVMAAAVTLRAPHGGIFVFFAIGNLLWFVIAVALGTVVS
. : : . . . .: .. . :.* :* .. :.: :. *.
Mvan_5589|M.vanbaalenii_PYR-1 KADDVARLREATTTEEVKAILMAADK
Mflv_1219|M.gilvum_PYR-GCK KPDEVARLRAAQTPEEIRSILMGS--
MSMEG_0085|M.smegmatis_MC2_155 ALTVIAAKQFTKRPVTAPEAPALATT
:* : : . :