For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQEVDTVERAAATPDHPQPYRELGLKDDEYERIREILGRRPTDAELAMYSVMWSEHCSYKSSKVHLRYFG ETTTEKMRETMLAGIGENAGVVDIGDGWAATFKVESHNHPSYIEPYQGAATGVGGIVRDIMAMGARPVAV MDQLRFGAADAPDTRRVLDGVVRGIGGYGNSLGLPNIGGETVFDASYAGNPLVNALCVGVLRKEDLKLAF ASGAGNKIILFGARTGLDGIGGVSVLASETFGGDESGAGRKKLPSVQVGDPFTEKVLIECCLELYAADLV VGIQDLGGAGLSCATSELASAGDGGMRVELERVPLRAANMTPAEILSSESQERMCAVVTPDNVDAFMAVC RKWDVLATDIGEVTDSGRLEITWHGETVVDVPPRTVAHEGPVYERPVQRPDSQDALVANTTASLERPSTG DELRATLLQLLGSPALCSRAFITEQYDRYVRGNTVLAEHADGGVLRVDEQTGRGIAISTDASGRYTALDP YAGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPGVMWQFSQAVRGLADGAAALGIPVTGGNVSFYN QTGTTAILPTPVVGVLGVIDDVKRRIPTGFGTEPGETLILLGDTRDEFDGSIWAQVTADHLGGVPPTVDL DRERLLAEVLTAASRDGLVSAAHDLSEGGLIQAVVEASLAGETGCRIVLPEGWNPFVTLFSESAGRVLVA VPRTEESRFTAMCEARGLPANRIGVVDQGPDGGEPAVEVQGLFTVTLEELRRTSEGVLPGLFG*
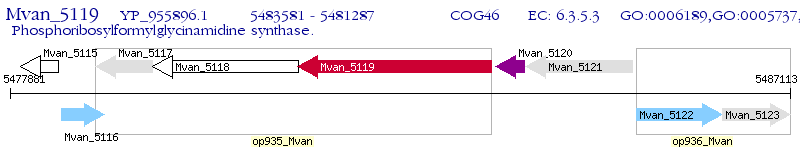
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5119 | - | - | 100% (764) | phosphoribosylformylglycinamidine synthase II |
| M. vanbaalenii PYR-1 | Mvan_2362 | - | 3e-05 | 27.27% (209) | hydrogenase expression/formation protein HypE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0826 | purL | 0.0 | 86.32% (760) | phosphoribosylformylglycinamidine synthase II |
| M. gilvum PYR-GCK | Mflv_1630 | - | 0.0 | 91.81% (781) | phosphoribosylformylglycinamidine synthase II |
| M. tuberculosis H37Rv | Rv0803 | purL | 0.0 | 86.32% (760) | phosphoribosylformylglycinamidine synthase II |
| M. leprae Br4923 | MLBr_02211 | purL | 0.0 | 83.42% (760) | phosphoribosylformylglycinamidine synthase II |
| M. abscessus ATCC 19977 | MAB_0707 | - | 0.0 | 87.06% (765) | phosphoribosylformylglycinamidine synthase II |
| M. marinum M | MMAR_4888 | purL | 0.0 | 84.69% (764) | phosphoribosylformylglycinamidine synthase II, PurL |
| M. avium 104 | MAV_0744 | purL | 0.0 | 86.63% (763) | phosphoribosylformylglycinamidine synthase II |
| M. smegmatis MC2 155 | MSMEG_5824 | purL | 0.0 | 88.63% (765) | phosphoribosylformylglycinamidine synthase II |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0410 | purL | 0.0 | 84.42% (764) | phosphoribosylformylglycinamidine synthase II |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0826|M.bovis_AF2122/97 -----------MLDTVEHAATTPDQPQPYGELGLKDDEYRRIRQILGRRP
Rv0803|M.tuberculosis_H37Rv -----------MLDTVEHAATTPDQPQPYGELGLKDDEYRRIRQILGRRP
MAV_0744|M.avium_104 MTVSPTSAPTQAIDTVERAATTPDEPQPFGELGLKDDEYQRIREILGRRP
MMAR_4888|M.marinum_M ----MTSARSHQTDTVAHAATTPDQPQPFHELGLKDDEYQRIRDILGRRP
MUL_0410|M.ulcerans_Agy99 ----MTSARSHQTDTVAHAATTPDQPQPFHELGLKDDEYQRIRDILGRRP
MLBr_02211|M.leprae_Br4923 -----------MIDTVEYAATTPDQPQPFAELGLREDEYQRVREILGRRP
Mvan_5119|M.vanbaalenii_PYR-1 --------MTQEVDTVERAAATPDHPQPYRELGLKDDEYERIREILGRRP
Mflv_1630|M.gilvum_PYR-GCK --------MTQEVDTVERAAATPGQPQPYRELGLKDDEYERIREILGRRP
MAB_0707|M.abscessus_ATCC_1997 --------MTSRVDTVDNAASTPDHPQPFAELGLKDDEYARIREILGRRP
MSMEG_5824|M.smegmatis_MC2_155 ----MTSELTHQTDTVERAAATPDQPQPYRELGLKDDEYQRIRDILGRRP
*** **:**..***: ****::*** *:*:******
Mb0826|M.bovis_AF2122/97 TDTELAMYSVMWSEHCSYKSSKVHLRYFGETTSDEMRAAMLAGIGENAGV
Rv0803|M.tuberculosis_H37Rv TDTELAMYSVMWSEHCSYKSSKVHLRYFGETTSDEMRAAMLAGIGENAGV
MAV_0744|M.avium_104 TDTELAMYSVMWSEHCSYKSSKVHLRYFGETTTDEMRAGMLAGIGENAGV
MMAR_4888|M.marinum_M TDTELAMYSVMWSEHCSYKSSKVHLRYFGETTTDEMRAGMLAGIGENAGV
MUL_0410|M.ulcerans_Agy99 TDTELAMYSVMWSEHCSYKSSKVHLRYFGETTTDEMRAGMLAGIGENAGV
MLBr_02211|M.leprae_Br4923 TDTELAMYSVMWSEHCSYKSSKVHLRYFGETTTEEMRTGMLAGIGENAGV
Mvan_5119|M.vanbaalenii_PYR-1 TDAELAMYSVMWSEHCSYKSSKVHLRYFGETTTEKMRETMLAGIGENAGV
Mflv_1630|M.gilvum_PYR-GCK TDAELAMYSVMWSEHCSYKSSKVHLRYFGETTTEKMREIMLAGIGENAGV
MAB_0707|M.abscessus_ATCC_1997 TDAELAMYSVMWSEHCSYKSSKVHLRYFGQTTTEEMRSAMLAGIGENAGV
MSMEG_5824|M.smegmatis_MC2_155 TDAELAMYSVMWSEHCSYKSSKVHLRYFGETTTEKMRAGMLAGIGENAGV
**:**************************:**:::** ***********
Mb0826|M.bovis_AF2122/97 VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPVAVM
Rv0803|M.tuberculosis_H37Rv VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPVAVM
MAV_0744|M.avium_104 VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPVAVM
MMAR_4888|M.marinum_M VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPIAVM
MUL_0410|M.ulcerans_Agy99 VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPIAVV
MLBr_02211|M.leprae_Br4923 VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPVAVM
Mvan_5119|M.vanbaalenii_PYR-1 VDIGDGWAATFKVESHNHPSYIEPYQGAATGVGGIVRDIMAMGARPVAVM
Mflv_1630|M.gilvum_PYR-GCK VDIGDGWAATFKVESHNHPSYIEPYQGAATGVGGIVRDIMAMGARPVAVM
MAB_0707|M.abscessus_ATCC_1997 VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPIAVM
MSMEG_5824|M.smegmatis_MC2_155 VDIGDGWAVTFKVESHNHPSYVEPYQGAATGVGGIVRDIMAMGARPVAVM
********.************:************************:**:
Mb0826|M.bovis_AF2122/97 DQLRFGAADAPDTRRVLDGVVRGIGGYGNSLGLPNIGGETVFDPCYAGNP
Rv0803|M.tuberculosis_H37Rv DQLRFGAADAPDTRRVLDGVVRGIGGYGNSLGLPNIGGETVFDPCYAGNP
MAV_0744|M.avium_104 DQLRFGAADAPDTRRVLDGVVRGIGGYGNSLGLPNIGGETVFDACYAGNP
MMAR_4888|M.marinum_M DQLRFGAADAPDTRRVLDGVVRGVGGYGNSLGLPNIGGETVFDACYAGNP
MUL_0410|M.ulcerans_Agy99 DQLRFGAADAPDTRRVLDGVVRGVGGYGNSLGLPNIGGDTVFDACYAGNP
MLBr_02211|M.leprae_Br4923 DQLRFGAADALDTRRVLDGVVRGIGGYGNSLGLPNIGGETVFDSCYDGNP
Mvan_5119|M.vanbaalenii_PYR-1 DQLRFGAADAPDTRRVLDGVVRGIGGYGNSLGLPNIGGETVFDASYAGNP
Mflv_1630|M.gilvum_PYR-GCK DQLRFGAADAPDTRRVLDGVVRGIGGYGNSLGLPNIGGETIFDASYAGNP
MAB_0707|M.abscessus_ATCC_1997 DQLRFGAADAPDTRRVFDGVVRGIGGYGNSLGLPNIGGETVFDASYAGNP
MSMEG_5824|M.smegmatis_MC2_155 DQLRFGPADAPDTRRVLDGVVRGVGGYGNSLGLPNIGGETVFDASYAGNP
******.*** *****:******:**************:*:**..* ***
Mb0826|M.bovis_AF2122/97 LVNALCVGVLRQEDLHLAFASGAGNKIILFGARTGLDGIGGVSVLASDTF
Rv0803|M.tuberculosis_H37Rv LVNALCVGVLRQEDLHLAFASGAGNKIILFGARTGLDGIGGVSVLASDTF
MAV_0744|M.avium_104 LVNAMCVGVLRQEDLHLAFASGAGNKIILFGARTGLDGIGGVSVLASDTF
MMAR_4888|M.marinum_M LVNALCVGVLRQEDLHLAFASGAGNKIILFGARTGLDGIGGVSVLASDTF
MUL_0410|M.ulcerans_Agy99 LVNALCVGVLRQEDLHLAFASGAGNKIILFGARTGLDGIGGVSVLASDTF
MLBr_02211|M.leprae_Br4923 LVNALCVGVLRQEDLHLAFASGAGNKIILFGACTGLDGIGGVSVLASDTF
Mvan_5119|M.vanbaalenii_PYR-1 LVNALCVGVLRKEDLKLAFASGAGNKIILFGARTGLDGIGGVSVLASETF
Mflv_1630|M.gilvum_PYR-GCK LVNALCVGVLRKEDLKLAFASGAGNKIILFGARTGLDGIGGVSVLASETF
MAB_0707|M.abscessus_ATCC_1997 LVNALCAGVLRKEDLHLAFASGAGNKIILFGARTGLDGIGGVSVLASETF
MSMEG_5824|M.smegmatis_MC2_155 LVNALCVGALRKEDLHLAFASGTGNKIILFGARTGLDGIGGVSVLASETF
****:*.*.**:***:******:********* **************:**
Mb0826|M.bovis_AF2122/97 DAEG----------------SRKKLPSVQVGDPFMEKVLIECCLELYAGG
Rv0803|M.tuberculosis_H37Rv DAEG----------------SRKKLPSVQVGDPFMEKVLIECCLELYAGG
MAV_0744|M.avium_104 DAEN----------------SRKKLPSVQVGDPFMEKVLIECCLELYAGG
MMAR_4888|M.marinum_M SSDGEGEG------------ARKKLPSVQVGDPFMEKVLIECCLELYSGG
MUL_0410|M.ulcerans_Agy99 SSDGEGEG------------ARKKLPSVQVGDPFMEKVLIECCLELYSGG
MLBr_02211|M.leprae_Br4923 DAEG----------------ARKKLPSVQVGDPFMEKVLIECCLELYAGG
Mvan_5119|M.vanbaalenii_PYR-1 GGDESGAG-------------RKKLPSVQVGDPFTEKVLIECCLELYAAD
Mflv_1630|M.gilvum_PYR-GCK GGDESGAGGHEAGGGAGPSRSRKKLPSVQVGDPFTEKVLIECCLELYAAD
MAB_0707|M.abscessus_ATCC_1997 GGDEADGAS------------RKKLPSVQVGDPFTEKVLIECCLELYAAH
MSMEG_5824|M.smegmatis_MC2_155 GGDEGSGPG------------RKKLPSVQVGDPFMEKVLIECCLELYSAG
..: ************* ************:.
Mb0826|M.bovis_AF2122/97 LVIGIQDLGGAGLSCATSELASAGDGGMTIQLDSVPLRAKEMTPAEVLCS
Rv0803|M.tuberculosis_H37Rv LVIGIQDLGGAGLSCATSELASAGDGGMTIQLDSVPLRAKEMTPAEVLCS
MAV_0744|M.avium_104 LVIGIQDLGGAGLACATSELASAGDGGMEVRLEAVPLRTAGMTPAEVLCS
MMAR_4888|M.marinum_M LVIGIQDLGGAGLSCATSELASAGDGGMAINLETVPLRAKEMTPAEVLCS
MUL_0410|M.ulcerans_Agy99 LVIGIQDLGGAGLSCATSELASAGDGGMAINLEAVPLRAKEMTPAEVLCS
MLBr_02211|M.leprae_Br4923 LVIGIQDLGGAGLSCATSELASAGDVGMAIQLDTVPRRAKDMTPAEVFCS
Mvan_5119|M.vanbaalenii_PYR-1 LVVGIQDLGGAGLSCATSELASAGDGGMRVELERVPLRAANMTPAEILSS
Mflv_1630|M.gilvum_PYR-GCK LVVGIQDLGGAGLSCATSELASAGDGGMRVELEQVPLRAANMTPAEILSS
MAB_0707|M.abscessus_ATCC_1997 LVVGIQDLGGAGLSCATSELASAGDGGMHIDLDKVPLRATGMTPAEVLSS
MSMEG_5824|M.smegmatis_MC2_155 LVVGIQDLGGAGLSCATSELASAGDGGMHIELDKVPLRAANMTPAEVLSS
**:**********:*********** ** : *: ** *: *****::.*
Mb0826|M.bovis_AF2122/97 ESQERMCAVVSPKNVDAFLAVCRKWEVLATVIGEVTDGDRLQITWHGETV
Rv0803|M.tuberculosis_H37Rv ESQERMCAVVSPKNVDAFLAVCRKWEVLATVIGEVTDGDRLQITWHGETV
MAV_0744|M.avium_104 ESQERMCAVVTPENVDAFLAVCRKWDVLATVIGEVTDGDRLRITWHGQTV
MMAR_4888|M.marinum_M ESQERMCAVVSPENVDAFMAVCRKWDVLATVIGEVTEGDRLRITWHGETV
MUL_0410|M.ulcerans_Agy99 ESQERMCAVVSPENVDAFMAVCRKWDVLATVIGEVTEGDRLRITWHGETV
MLBr_02211|M.leprae_Br4923 ESQERMCAVVAPENVDAFLAVCRKWEVLATVIGEVTDGDRLRITWHGETV
Mvan_5119|M.vanbaalenii_PYR-1 ESQERMCAVVTPDNVDAFMAVCRKWDVLATDIGEVTDSGRLEITWHGETV
Mflv_1630|M.gilvum_PYR-GCK ESQERMCAVVTPENVDAFMEVCRKWDVLATVIGEVTDSGRLEITWHGETV
MAB_0707|M.abscessus_ATCC_1997 ESQERMCAVVTPENVDAFMAVCRKWDVLATVIGEVTDGDRLRITWHGETV
MSMEG_5824|M.smegmatis_MC2_155 ESQERMCAVVTPENVDAFLAVCRKWEVLATVIGEVTDGDRLQITWHGETV
**********:*.*****: *****:**** *****:..**.*****:**
Mb0826|M.bovis_AF2122/97 VDVPPRTVAHEGPVYQRPVARPDTQDALNADRSAKLSRPVTGDELRATLL
Rv0803|M.tuberculosis_H37Rv VDVPPRTVAHEGPVYQRPVARPDTQDALNADRSAKLSRPVTGDELRATLL
MAV_0744|M.avium_104 VDVPPRTVAHEGPVYQRPLARPDTQDALNADSSARLPRPATGAELRATLL
MMAR_4888|M.marinum_M VDVPPRTVAHEGPVYQRPVARPETQDALNADSSKSLPRPATGDELRATLL
MUL_0410|M.ulcerans_Agy99 VDVPPRTVAHEGPVYQRPVARPDTQDALNADSSKSLPRPATGDELRATLL
MLBr_02211|M.leprae_Br4923 VDVPPRTVAHEGPVYQRPVSRPESQEALNADSSKGLPRPVSGDELRATLL
Mvan_5119|M.vanbaalenii_PYR-1 VDVPPRTVAHEGPVYERPVQRPDSQDALVANTTASLERPSTGDELRATLL
Mflv_1630|M.gilvum_PYR-GCK VDVPPRTVAHEGPVYERPVQRPDTQDALIADTSASLPRPATGEELRATLL
MAB_0707|M.abscessus_ATCC_1997 VDVPPRTVAHEGPVYQRPVARPDTQDALIADTAGGLARPASAAELRQTLL
MSMEG_5824|M.smegmatis_MC2_155 VDVPPRTVAHEGPVYERPVERPECQDALIADTSAKLPRPTTGDELKETLL
***************:**: **: *:** *: : * ** :. **: ***
Mb0826|M.bovis_AF2122/97 ALLGSPHLCSRAFITEQYDRYVRGNTVLAEHADGGMLRIDESTGRGIAVS
Rv0803|M.tuberculosis_H37Rv ALLGSPHLCSRAFITEQYDRYVRGNTVLAEHADGGMLRIDESTGRGIAVS
MAV_0744|M.avium_104 ALLGSPHLCSRAFITEQYDRYVRGNTVLAEHADGGVLRVDETTGRGIAVS
MMAR_4888|M.marinum_M ALLGSPHLCSRAFITEQYDRYVRGNTVLAEHADGGVLRIDEATGRGIALS
MUL_0410|M.ulcerans_Agy99 ALLGSPHLCSRAFITEQYDRYVRGNTVLAEHANGGVLRIDEATGRGIALS
MLBr_02211|M.leprae_Br4923 ALLGSPHLCSRAFITEQYDRYVRGNTVLAEHADAGVLRIDESTGRGIALS
Mvan_5119|M.vanbaalenii_PYR-1 QLLGSPALCSRAFITEQYDRYVRGNTVLAEHADGGVLRVDEQTGRGIAIS
Mflv_1630|M.gilvum_PYR-GCK AMVGSPHLCSRAFITEQYDRYVRGNTVLAEYADGGVLRVDEQTGRGIAIS
MAB_0707|M.abscessus_ATCC_1997 DMIGSPHLCSRAFITEQYDRYVRGNTVLAEHADSGVIRVDEQTGRGIALA
MSMEG_5824|M.smegmatis_MC2_155 KLLGSPHLCSRAFITEQYDRYVRGNTVLAEHADGGVLRIDENTGRGIAVS
::*** ***********************:*:.*::*:** ******::
Mb0826|M.bovis_AF2122/97 TDASGRYTLLDPYAGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPG
Rv0803|M.tuberculosis_H37Rv TDASGRYTLLDPYAGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPG
MAV_0744|M.avium_104 TDASGRYTMLDPYAGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPA
MMAR_4888|M.marinum_M TDASGRYTLLDPYTGAQLALAEAYRNVAATGATPVAVTNCLNFGSPEDPG
MUL_0410|M.ulcerans_Agy99 TDASGRYTLLDPYTGAQLALAEAYRNVAATGATPVAVTNCLNFGSPEDPG
MLBr_02211|M.leprae_Br4923 TDASGRYTRLDPYAGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPG
Mvan_5119|M.vanbaalenii_PYR-1 TDASGRYTALDPYAGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPG
Mflv_1630|M.gilvum_PYR-GCK TDASGRYTALDPYAGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPG
MAB_0707|M.abscessus_ATCC_1997 TDASGRYTLLDPYRGAQLALAEAYRNVAASGATPVAVTNCLNFGSPEDPG
MSMEG_5824|M.smegmatis_MC2_155 TDASGRYTQLDPYTGAQLALAEAYRNVAVTGATPVAVTNCLNFGSPEDPG
******** **** **************.:*******************.
Mb0826|M.bovis_AF2122/97 VMWQFTQAVRGLADGCADLGIPVTGGNVSFYNQTGSAAILPTPVVGVLGV
Rv0803|M.tuberculosis_H37Rv VMWQFTQAVRGLADGCADLGIPVTGGNVSFYNQTGSAAILPTPVVGVLGV
MAV_0744|M.avium_104 VMWQFAQAVRGLADGCVALGIPVTGGNVSFYNQTGSAAILPTPVVGVLGV
MMAR_4888|M.marinum_M VMWQFAQAVRGLADGCVALGIPVTGGNVSFYNQTGSTAIMPTPVVGVLGV
MUL_0410|M.ulcerans_Agy99 VMWQFAQAVRGLADGCVALGIPVTGGNVSFYNQTGSTAIMPTPVVGVLGV
MLBr_02211|M.leprae_Br4923 VMWQFAQAVRGLADGCAALKIPVTGGNVSFYNQTGAVAILPTPVVGVLGV
Mvan_5119|M.vanbaalenii_PYR-1 VMWQFSQAVRGLADGAAALGIPVTGGNVSFYNQTGTTAILPTPVVGVLGV
Mflv_1630|M.gilvum_PYR-GCK VMWQFSQAVRGLADGAAALGIPVTGGNVSFYNQTGTTAILPTPVVGVLGI
MAB_0707|M.abscessus_ATCC_1997 VMWQFSEAVRGLADGCVTLGIPVTGGNVSFYNQTGTTPILPTPVVGVLGV
MSMEG_5824|M.smegmatis_MC2_155 VMWQFSQAVRGLADGCAALGIPVTGGNVSFYNQTGSTPILPTPVVGVLGV
*****::********.. * ***************:..*:*********:
Mb0826|M.bovis_AF2122/97 IDDVRRRIPTGLGAEPGETLMLLGDTRDEFDGSVWAQVTADHLGGLPPVV
Rv0803|M.tuberculosis_H37Rv IDDVRRRIPTGLGAEPGETLMLLGDTRDEFDGSVWAQVTADHLGGLPPVV
MAV_0744|M.avium_104 LDDVSRRLPTALGAEPGETLMLLGETRDEFDGSVWAQVTADHLGGLPPKV
MMAR_4888|M.marinum_M IDDVTRRIPTAMGAEPSETLMLLGDTRDELDGSIWAQVTGEHLGGTPPAV
MUL_0410|M.ulcerans_Agy99 IDDVTRRIPTAMGAEPSETLMLLGDTRDELDGSIWAQVTGEHLGGTPPAV
MLBr_02211|M.leprae_Br4923 LDNVARRIHTSLGTEPGEILMLLGDTYDEFDGSVWAQVMAGHLGGLPPMV
Mvan_5119|M.vanbaalenii_PYR-1 IDDVKRRIPTGFGTEPGETLILLGDTRDEFDGSIWAQVTADHLGGVPPTV
Mflv_1630|M.gilvum_PYR-GCK LDNVSRRLPTAFGSEPGETLILLGDTHDEFDGSIWAEVTAGHLGGVPPKV
MAB_0707|M.abscessus_ATCC_1997 IDDVNRRIPTGFGTEPGETLILLGDTADEFDGSIWAQVAHDHLGGTPPKV
MSMEG_5824|M.smegmatis_MC2_155 IDDVKRRIPTGLGTEPGETLLLLGDTHDEFDGSIWAQVTADHLGGVPPKV
:*:* **: *.:*:**.* *:***:* **:***:**:* **** ** *
Mb0826|M.bovis_AF2122/97 DLAREKLLAAVLSSASRDGLVSAAHDLSEGGLAQAIVESALAGETGCRIV
Rv0803|M.tuberculosis_H37Rv DLAREKLLAAVLSSASRDGLVSAAHDLSEGGLAQAIVESALAGETGCRIV
MAV_0744|M.avium_104 DLAREKLLAEVLRAASRDGLVSAAHDLSEGGLAQAVVEAALAGETGCRIV
MMAR_4888|M.marinum_M DLAREKLLADVLRAASRDGLVSAAHDLSEGGLAQAIVESALAGETGCRIV
MUL_0410|M.ulcerans_Agy99 DLAREKLLADVLRAASRDGLVSAAHDLSEGGLAQAIVESALAGETGCRIV
MLBr_02211|M.leprae_Br4923 DLAREKLLAEVLSSASRDELVSAAHDLSEGGLAQAIVESALAGETGCRIA
Mvan_5119|M.vanbaalenii_PYR-1 DLDRERLLAEVLTAASRDGLVSAAHDLSEGGLIQAVVEASLAGETGCRIV
Mflv_1630|M.gilvum_PYR-GCK DLEREQLLAEVLTAASRDGLVSAAHDVSEGGLIQAVVEAALAGETGCRIV
MAB_0707|M.abscessus_ATCC_1997 DLAREQLLAQVLTAASRDGLVSAAHDLSEGGLIQAVVESALAGETGCRLL
MSMEG_5824|M.smegmatis_MC2_155 DLDREKLLAEVLTAASRDGLISAAHDLAEGGLIQAVVESALAGETGCRVI
** **:*** ** :**** *:*****::**** **:**::********:
Mb0826|M.bovis_AF2122/97 LPEGADP----FVLLFSESAGRVLVAVPRTEESRFRGMCEARGLPAVRIG
Rv0803|M.tuberculosis_H37Rv LPEGADP----FVLLFSESAGRVLVAVPRTEESRFRGMCEARGLPAVRIG
MAV_0744|M.avium_104 LPEDADP----FVTLFSESAGRVLVAVPRTEESRFRSMCEARGLPAVRIG
MMAR_4888|M.marinum_M LPDDADP----FVMLFSESAGRVLVAVPRTEESRLRAMCAARGLPAVRIG
MUL_0410|M.ulcerans_Agy99 LPDDADP----FVMLFSESAGRVLVAVPRTEESRLRAMCEARRLPAVRIG
MLBr_02211|M.leprae_Br4923 LPEDADP----FVMLFSESAGRVLVAVPRPEESRFRSMCEARGLPAMRIG
Mvan_5119|M.vanbaalenii_PYR-1 LPEGWNP----FVTLFSESAGRVLVAVPRTEESRFTAMCEARGLPANRIG
Mflv_1630|M.gilvum_PYR-GCK LPEGFHDGTGVFVFLFSESSGRVLVAVPRTEESRFTAMCQARGLPATRIG
MAB_0707|M.abscessus_ATCC_1997 LPEGADP----FVALFSESAGRVLVAVPRTEESRFMSMCEARQLPAVRIG
MSMEG_5824|M.smegmatis_MC2_155 IPEGADP----FVFLFSESSGRVLVAVPRTEESRFRSMCEARGLPVTRVG
:*:. . ** *****:*********.****: .** ** **. *:*
Mb0826|M.bovis_AF2122/97 VVDQGSD----AVEVQGLFAVSLAELRATSEAVLPRYFG
Rv0803|M.tuberculosis_H37Rv VVDQGSD----AVEVQGLFAVSLAELRATSEAVLPRYFG
MAV_0744|M.avium_104 VVDQASD----EVEVQGLFTVSLAELRQTSESVLPRLFG
MMAR_4888|M.marinum_M VVDQASD----DIEVQGLFTVSLAELRATSEAVLPRFFG
MUL_0410|M.ulcerans_Agy99 VVDQASD----DIEVQGLFTVSLAELRATSEAVLPRFFG
MLBr_02211|M.leprae_Br4923 VVDQGSD----SIEVRGQFTVSLAELRMTFEAVLPRFFG
Mvan_5119|M.vanbaalenii_PYR-1 VVDQGPDGGEPAVEVQGLFTVTLEELRRTSEGVLPGLFG
Mflv_1630|M.gilvum_PYR-GCK VVDQGPEGGEPTVEVQGQFTVGIEELRRTSEGVLPGLFG
MAB_0707|M.abscessus_ATCC_1997 VVDQGSD----SVEVQGQFSVTLAELREIHEGVLPGLFG
MSMEG_5824|M.smegmatis_MC2_155 VVDQGSD----SVEVQDQFSVALSELRSTSEGVLPRLFG
****..: :**:. *:* : *** *.*** **