For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MATPAAQELDPLALERQVCFALAATNRAVLAIYRPLLEPLGLTHPQYLVMLSLWDQQKRSSGRPSPMSVK QIARTLQIDSATLSPMLKRLEAIGLITRARNTADERSTDVQLTDAGAELRRRALDIPAAVIERLGVDVAE LEHLHEVLTRINSAAVAAGALQS
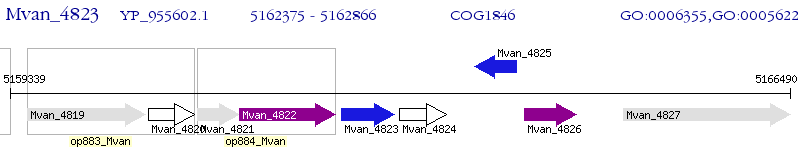
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4823 | - | - | 100% (163) | MarR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1524 | - | 9e-26 | 41.10% (146) | MarR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_6049 | - | 1e-05 | 31.33% (83) | MarR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2911 | - | 3e-05 | 28.70% (108) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_1910 | - | 8e-68 | 81.65% (158) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2887 | - | 3e-05 | 28.70% (108) | transcriptional regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1029c | - | 1e-58 | 69.14% (162) | MarR family transcriptional regulator |
| M. marinum M | MMAR_0145 | - | 5e-63 | 73.62% (163) | transcriptional regulator |
| M. avium 104 | MAV_3741 | - | 4e-05 | 31.03% (87) | MarR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0538 | - | 5e-62 | 71.43% (161) | regulatory protein, MarR |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4952 | - | 4e-63 | 73.62% (163) | transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4823|M.vanbaalenii_PYR-1 --------------------------MATPAAQELDPLALERQVCFALAA
Mflv_1910|M.gilvum_PYR-GCK ----------------------------MTVAAEADPLALERQVCFALAV
MMAR_0145|M.marinum_M --------------------------MAAPTTPQADPLALEQQVCFALAI
MUL_4952|M.ulcerans_Agy99 --------------------------MAAPTTPQADPLALEQQVCFALAI
MSMEG_0538|M.smegmatis_MC2_155 --------------------------MASPSSVDVDPLALERQVCFALAV
MAB_1029c|M.abscessus_ATCC_199 ---------------------------MTAVSPEVDPLALERQVCFALAV
Mb2911|M.bovis_AF2122/97 -----------------------------------MGLADDAPLGYLLYR
Rv2887|M.tuberculosis_H37Rv -----------------------------------MGLADDAPLGYLLYR
MAV_3741|M.avium_104 MPPDWLPPDGYPVAGPAITPPNTRTGAPRYVKIVDMTEADDAPLGYLLYR
* : : : *
Mvan_4823|M.vanbaalenii_PYR-1 TNRAVLAIYRPLLEPLGLTHPQYLVMLSLWDQQKRSSGRP-SPMSVKQIA
Mflv_1910|M.gilvum_PYR-GCK TNRAVLAIYRPLLEPLGLTHPQYLVMLALWDHHKKASGQA-TPLSVKKIA
MMAR_0145|M.marinum_M TNRAVLAVYRPLLEPLGLTHPQYLVMLALWDNQRTSSGQG-SALAVKQIA
MUL_4952|M.ulcerans_Agy99 TNRAVLAVYRPLLEPLGLTHPQYLVMLALWDNQRTSSGQG-SALAVKQIA
MSMEG_0538|M.smegmatis_MC2_155 TNRAVLSIYRPLLEPLGLTHPQYLVMLALWDHHKIRSAGV-DPLSVKQIA
MAB_1029c|M.abscessus_ATCC_199 TNRAVLAVYRPLLAPLGITHPQYLVMLTLWDHAKSGADDTREALSVKQIA
Mb2911|M.bovis_AF2122/97 VGAVLRPEVSAALSPLGLTLPEFVCLRMLSQS---------PGLSSAELA
Rv2887|M.tuberculosis_H37Rv VGAVLRPEVSAALSPLGLTLPEFVCLRMLSQS---------PGLSSAELA
MAV_3741|M.avium_104 VGAALRPEVSGALGPLGLTLPEFVCLRILSMF---------PGMSSAELS
.. .: . * ***:* *::: : * :: :::
Mvan_4823|M.vanbaalenii_PYR-1 RTLQIDSATLSPMLKRLEAIGLITRARNTADERSTDVQLTDAGAELRRRA
Mflv_1910|M.gilvum_PYR-GCK TTLQMDSATLSPMLKRLEALGFISRTRNAADERSTDVQLTDAGVALRTRA
MMAR_0145|M.marinum_M ATLQLDSATLSPMLKRLEALGLVTRTRNATDERSTDVKLTRFGIALRERA
MUL_4952|M.ulcerans_Agy99 ATLQLDSATLSPMLKRLEALGLVTRTRNATDERSTDVKLTRFGIALRERA
MSMEG_0538|M.smegmatis_MC2_155 ATLQLDSATLSPMLKRLEALGLITRTRSVGDERATDIRLTEAGIALRERA
MAB_1029c|M.abscessus_ATCC_199 ALLQTDSATISPMLKRLESLGLITRPRSTADERTTHVTLTSRGAALREQA
Mb2911|M.bovis_AF2122/97 RHASVTPQAMNTVLRKLEDAGAVARPASVSSGRSLPATLTARGRALAKRA
Rv2887|M.tuberculosis_H37Rv RHASVTPQAMNTVLRKLEDAGAVARPASVSSGRSLPATLTARGRALAKRA
MAV_3741|M.avium_104 RRAGVTPQAMNTVLRRLQEVGAVARPSSVSSGRSLPAHLTGAGRTLLKRA
. ::..:*::*: * ::*. .. . *: ** * * :*
Mvan_4823|M.vanbaalenii_PYR-1 LDIPAAVIERLGVDVAELEH--LHEVLTRINSAAVAAGALQS-
Mflv_1910|M.gilvum_PYR-GCK LDIPPAVIERLGVDLAELEH--LHHVLTRVNSA--AAGALQS-
MMAR_0145|M.marinum_M AEVPAAVIARLGVDPTELEQ--LHRVLTRINVAALAADAPQS-
MUL_4952|M.ulcerans_Agy99 AEVPAAVIARLGVDPTELEQ--LHRVLTRINVAALAADAPQS-
MSMEG_0538|M.smegmatis_MC2_155 LAIPPKVVERLGVDMAELEE--LHRVLTRVNTAALAANGVNH-
MAB_1029c|M.abscessus_ATCC_199 LSVPAAVVERLGVNLAELEN--LHKVLTRVNTAALAAGALDTE
Mb2911|M.bovis_AF2122/97 EAVVRAADARVLARLTAPQQREFKRMLEKLGSD----------
Rv2887|M.tuberculosis_H37Rv EAVVRAADARVLARLTAPQQREFKRMLEKLGSD----------
MAV_3741|M.avium_104 EAAVRGADARILAKLTETQQREFKRMLQKLGSDG---------
. *: . : :. ::.:* ::.