For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TGIGGHPGIPAEPKKRPAWMVGGLTVLSFVVLLWVIELFDSLTGHRLDGNGIRPLETDGLAGILFAPLLH SNWEHLLANTVPALVLGFLMTLAGLSRFIFATAIVWIVGGVGTWLIGNIGAHCPYVGVRCETNHIGASGL IFGWLAFLIVFGFFTRKVWEIVVGVVVLFVYGSVLLGVLPGTPGVSWQGHLSGAVAGVLAAYLLSGPERK ARERRKATNPYLTT*
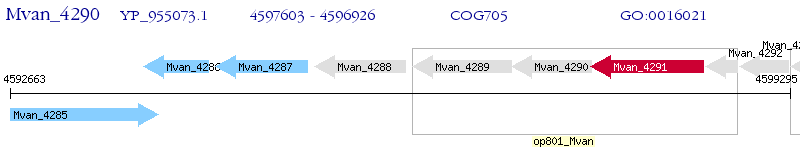
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4290 | - | - | 100% (225) | rhomboid family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1372 | - | 1e-83 | 66.22% (222) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_2355 | - | 1e-101 | 79.64% (221) | rhomboid family protein |
| M. tuberculosis H37Rv | Rv1337 | - | 1e-83 | 66.22% (222) | integral membrane protein |
| M. leprae Br4923 | MLBr_01171 | - | 8e-71 | 61.72% (209) | hypothetical protein MLBr_01171 |
| M. abscessus ATCC 19977 | MAB_1481 | - | 6e-81 | 67.57% (222) | hypothetical protein MAB_1481 |
| M. marinum M | MMAR_4059 | - | 3e-83 | 67.59% (216) | integral membrane protein |
| M. avium 104 | MAV_1554 | - | 1e-73 | 63.16% (209) | rhomboid family protein |
| M. smegmatis MC2 155 | MSMEG_4904 | - | 6e-86 | 68.02% (222) | rhomboid family protein |
| M. thermoresistible (build 8) | TH_1018 | - | 2e-83 | 72.14% (201) | PROBABLE INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4822 | - | 6e-05 | 26.60% (203) | serine protease |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4290|M.vanbaalenii_PYR-1 ---------------MTGIGGHPGIPAEPKKR------PAWMVGGLTVLS
Mflv_2355|M.gilvum_PYR-GCK ---------------MTGTGGYPLQPAGPAKR------PVWVVGGVTVVS
MAB_1481|M.abscessus_ATCC_1997 ---------------MTGYGGGPDYPGSPSSRRERSGRPQWVTGGVLVAS
TH_1018|M.thermoresistible__bu -----------------------------------------VTGGLTILT
MLBr_01171|M.leprae_Br4923 MGA-PSACGGGSALMTLPEKNLGYKPETETNRR-----LRWMVGGVTILT
MAV_1554|M.avium_104 MG-----------------KTTGRAPAAQTKKR-----STAVAGATTIVT
Mb1372|M.bovis_AF2122/97 MGMTPRRKRRGGAVQITRPTGRPRTPTTQTTKR-----PRWVVGGTTILT
Rv1337|M.tuberculosis_H37Rv MGMTPRRKRRGGAVQITRPTGRPRTPTTQTTKR-----PRWVVGGTTILT
MMAR_4059|M.marinum_M ---------------MAR---HPRTPATQPEER-----PRWVTGGATILT
MSMEG_4904|M.smegmatis_MC2_155 -------------------MTLPTGPAPKKNKS-----PAWKTGGITILT
MUL_4822|M.ulcerans_Agy99 --MRSGAVGHQCVECVRADARTVRQPRTQFGGR------QRSVTPVVTFS
. :
Mvan_4290|M.vanbaalenii_PYR-1 FVVLLWVIELFDSLTGHRLDGNGIRP--LETDGLAGILFAPLLHSNWEHL
Mflv_2355|M.gilvum_PYR-GCK FVVLLWLIELFDTLTGHRLDDNGIRP--LETDGLTGILVAPLLHSNWEHL
MAB_1481|M.abscessus_ATCC_1997 FVALLYVVEGIDSVTGHRLDENGIRP--LETDGLDGILFAPLLHANWGHL
TH_1018|M.thermoresistible__bu FVALLYVIEAYDAVTGNQLDQNGIRP--LDTDGLTGVLLAPLLHGGWEHL
MLBr_01171|M.leprae_Br4923 FMALLYLVELIDQLTRHSLDNNGIRL--LKTDVLWGISFAPVLHANWQHL
MAV_1554|M.avium_104 FVALLYLVELIDQLTRHALDVHGIRP--QRLDGLWGILFAPVLHESWQHL
Mb1372|M.bovis_AF2122/97 FVALLYLVELIDQLSGSRLDVNGIRP--LKTDGLWGVIFAPLLHANWHHL
Rv1337|M.tuberculosis_H37Rv FVALLYLVELIDQLSGSRLDVNGIRP--LKTDGLWGVIFAPLLHANWHHL
MMAR_4059|M.marinum_M FVALLYLVELIDQLTRHSLDLNGIRP--LQTDGLWGIIFAPLLHANWAHL
MSMEG_4904|M.smegmatis_MC2_155 FVALLYVVELADQLSGGRLDRNGIRP--LETDGLWGVLFAPLLHANWEHL
MUL_4822|M.ulcerans_Agy99 LIAVSVLVFVLQLVVAGLGPHLALWPPAVADGQLYRLVTSAFMHYGRLHL
::.: :: : : .: . * : :..:* . **
Mvan_4290|M.vanbaalenii_PYR-1 LANTVPALVLG-FLMTLAGLSRFIFATAIVWIVGGVGTWLIGNIGAHCPY
Mflv_2355|M.gilvum_PYR-GCK IANTGPALVLG-FLMTLAGLSRFIFATAIIWFAGGLGTWLIGNVGAPT-Y
MAB_1481|M.abscessus_ATCC_1997 VANTVPALVLG-FLAAAAGVGRFLLATAIIWLLGGVGTWLIGNIGLPY--
TH_1018|M.thermoresistible__bu MANTGPALVLG-FLMTLAGMSRFIFATAIIWILGGLGTWLIGDLGAPAG-
MLBr_01171|M.leprae_Br4923 VANTIPLLVLG-FLIALAGLSRFIWVTAMVWIFGGSATWLIGNMGSSFG-
MAV_1554|M.avium_104 MANTVPLLVLG-FLMTLAGLARFVWATVIIWIVGGFGTWLIGNVGSSCG-
Mb1372|M.bovis_AF2122/97 MANTIPLLVLG-FLMTLAGLSRFVWATAIIWILGGLGTWLIGNVGSSCG-
Rv1337|M.tuberculosis_H37Rv MANTIPLLVLG-FLMTLAGLSRFVWATAIIWILGGLGTWLIGNVGSSCG-
MMAR_4059|M.marinum_M MANTVPLLVLG-FLMTLAGLARFVWATAIVWILGGFGTWLIGNVGSSCG-
MSMEG_4904|M.smegmatis_MC2_155 MANTVPALVLG-FLVTLAGLARFVWATAIIWILGGIGTWLIGDVGGCPL-
MUL_4822|M.ulcerans_Agy99 VFNMWALYVVGPPLEMWLGRLRFGGLYALSALGGSVLVYLIAPLNTATA-
: * . *:* * * ** .: : *. .:**. :.
Mvan_4290|M.vanbaalenii_PYR-1 VGVRCETNHIGASGLIFGWLAFLIVFGFFTRKVWEIVVGVVVLFVYGSVL
Mflv_2355|M.gilvum_PYR-GCK NGVIVETNHIGASGLIFGWLTFLIVFGFFTRTVWEIVVGVVVFFIYGGIL
MAB_1481|M.abscessus_ATCC_1997 -----ETNHIGASGLIFGWLTYVIVRGFFNRSVGQILIGAVVLVLYGGVL
TH_1018|M.thermoresistible__bu ----VSTVHIGASGLIFGWLTFLIVFGFFTRKVWQITVGIMVLLIYGSIL
MLBr_01171|M.leprae_Br4923 -----PTDHIGVSGLIFGWLAFLLVFGLFVRRGWD-IIGCMVLFAYGGVL
MAV_1554|M.avium_104 -----PTDHIGASGLIFGWLAFLLVFGIFVRRVRDIIIGLAVMFAYGGVL
Mb1372|M.bovis_AF2122/97 -----PTDHIGASGLIFGWLAFLLVFGLFVRKGWDIVIGLVVLFVYGGIL
Rv1337|M.tuberculosis_H37Rv -----PTDHIGASGLIFGWLAFLLVFGLFVRKGWDIVIGLVVLFVYGGIL
MMAR_4059|M.marinum_M -----PTDHIGASGLIFGWLAFLLVFGLFVRKVWDIIIGLAVLFFYGGIL
MSMEG_4904|M.smegmatis_MC2_155 -----PTNHIGASGLIFGWLTFLLVFGWFTRNAWQIVVGILVLLVYGGIL
MUL_4822|M.ulcerans_Agy99 ----------GASGAIFGLFGATFVVAKRLNLDIRWVVALIAINLAITFV
*.** *** : :* . . :. .: .:
Mvan_4290|M.vanbaalenii_PYR-1 LGVLP--GTPG-VSWQGHLSGAVAG--VLAAYLLSGPERK----------
Mflv_2355|M.gilvum_PYR-GCK LGVLP--GTFG-VSWQGHLCGALAG--VLAAYLLSGPERK----------
MAB_1481|M.abscessus_ATCC_1997 WGVLP--GQFG-VSWQGHLSGAVAG--VLAAYWLSGRERK----------
TH_1018|M.thermoresistible__bu LGVLP--GTFG-VSWQGHLCGAIAG--VIAAYVLSGPERR----------
MLBr_01171|M.leprae_Br4923 LGVMPVLGRCGGVSWQGHLCGAISG--VVAAYLLSAPERK----------
MAV_1554|M.avium_104 LGAMPVLGLCGGVSWQGHLCGAIAG--VLAAYLLSEPERK----------
Mb1372|M.bovis_AF2122/97 LGAMPVLGQCGGVSWQGHLSGAVAG--VVAAYLLSAPERK----------
Rv1337|M.tuberculosis_H37Rv LGAMPVLGQCGGVSWQGHLSGAVAG--VVAAYLLSAPERK----------
MMAR_4059|M.marinum_M LGALPRLGVCGGVSWQGHLCGALAG--VVAAYFLSAPERK----------
MSMEG_4904|M.smegmatis_MC2_155 WGAVPVLNGCGGVSWQGHLCGAIAG--VVAAYVLAGPERK----------
MUL_4822|M.ulcerans_Agy99 APAVG----SQLISWQGHVGGLITGALVAAAYVYAPRERRNFIQGAVTLT
.: :*****: * ::* * *** : **:
Mvan_4290|M.vanbaalenii_PYR-1 ----------ARERR-KATN-PYLTT-
Mflv_2355|M.gilvum_PYR-GCK ----------TRALR-RPVRPPSLNT-
MAB_1481|M.abscessus_ATCC_1997 ----------VQAAR-GPGVPPRLTP-
TH_1018|M.thermoresistible__bu ----------ARALR-RPVQPPRLTR-
MLBr_01171|M.leprae_Br4923 ----------TRALKEAGTDSPRLKT-
MAV_1554|M.avium_104 ----------GRSGRKTGAVAPRPKT-
Mb1372|M.bovis_AF2122/97 ----------ARALKRAGARSGHPKL-
Rv1337|M.tuberculosis_H37Rv ----------ARALKRAGARSGHPKL-
MMAR_4059|M.marinum_M ----------ARELKKAGERPSSPKT-
MSMEG_4904|M.smegmatis_MC2_155 ----------ARDSRKRSR--PYLTS-
MUL_4822|M.ulcerans_Agy99 VAVAFVVLIWWRTSELLAMFGGHLNLG
: . .