For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QNAELTFRSLGGASTVTGSKHLLESDGTRILVDCGLFQGVKNLRELNWAPLAVDPSSIDAVVVTHAHLDH TGYLPRLVRDGFRGPIVSTEATAAVADIILRDSAKLQERDAEFLNRHKATKHHPALPLYDSDDAQRALEL FDAHPFGEEFNLPGRGPAVTFRRAGHILGAATVDLAWHGRRIVFTGDLGRYDDPVMLDPEPVPAADYLVM ESTYGDRLHGHADPAEALAEVIDATVDRGGTVVIPAFAVGRAQALLYYLWRLHSAGRLPGIPIYLDSPMA INASDLLGAFPRDHRLPPEVYDDMCALATYTRDTDESKKITASREPKIVISASGMATGGRILHHLKAFAP DPRNTIIVTGYQVPGTRGRSIAAGEPYVKIHGEWIPINAQVANLQMLSAHADADELIRWAGGFVTAPRQV FVVHGEPQAADTLRRRLDREFGWQATVPRPNQQFSL*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
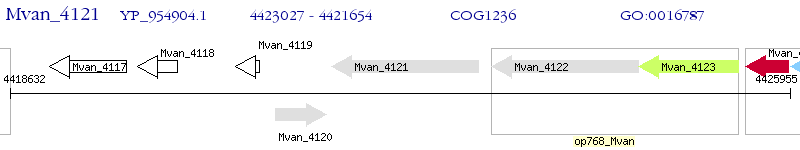
| M. vanbaalenii PYR-1 | Mvan_4121 | - | - | 100% (457) | beta-lactamase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_2388 | - | 2e-05 | 21.64% (439) | beta-lactamase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3999 | - | 4e-05 | 29.59% (98) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01512 | - | 2e-05 | 22.37% (447) | hypothetical protein MLBr_01512 |
| M. abscessus ATCC 19977 | MAB_3083c | - | 0.0001 | 28.57% (105) | hypothetical protein MAB_3083c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2685 | - | 6e-07 | 24.31% (436) | metallo-beta-lactamase superfamily protein |
| M. thermoresistible (build 8) | TH_0833 | - | 2e-09 | 22.97% (444) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3999|M.gilvum_PYR-GCK MDLTHTPP-----GPLAPGGLRVTALGGIGEIGRNMTVFEHLGRLLIVDC
MSMEG_2685|M.smegmatis_MC2_155 MSAELAPP-----PPLAPGGLRVTALGGISEIGRNMTVFEHLGRLLIVDC
TH_0833|M.thermoresistible__bu MSVPIPPPDLGPPGPLERGGLRVTALGGIGEIGRNMTVFEHRGRLLIIDC
MLBr_01512|M.leprae_Br4923 MDVALAPP-----GPLASGGLRVTALGGISEIGRNMTIFEHLGRLLIIDC
MAB_3083c|M.abscessus_ATCC_199 MAVTPPED-LRAPGPLADGALRVTALGGISEIGRNMTVFELRGRLLIIDC
Mvan_4121|M.vanbaalenii_PYR-1 ---------------MQNAELTFRSLGGASTVTGSKHLLESDGTRILVDC
: . * . :*** . : . ::* * :::**
Mflv_3999|M.gilvum_PYR-GCK GVLFPTHDEPGVDLILPDLRHVQDRLDDVEAIVVTHAHEDHIGAIPFVLK
MSMEG_2685|M.smegmatis_MC2_155 GVLFPGHDEPGVDLILPDLRHIEDRLDEIEALVVTHAHEDHIGAIPFLLK
TH_0833|M.thermoresistible__bu GVLFPGHDEPGVDLILPDLRHIENRLDDVEALLLTHAHEDHIGAVPFLLK
MLBr_01512|M.leprae_Br4923 GVLSPGHDEPGVDLILPDLRHIEDRLDDIEALVLTHAHEDHIGAIPFLLK
MAB_3083c|M.abscessus_ATCC_199 GVLFPGHNEPGVDLILPDIRHIEDRLEDVEALVLTHAHEDHIGAIPFLLK
Mvan_4121|M.vanbaalenii_PYR-1 GLFQGVKN-----LRELNWAPLAVDPSSIDAVVVTHAHLDHTGYLPRLVR
*:: :: * : : ..::*:::**** ** * :* :::
Mflv_3999|M.gilvum_PYR-GCK QRADIPIVGSKFTLALIAEKCR-EHRLSPKLVEVTEGTKSTHGVFECQYF
MSMEG_2685|M.smegmatis_MC2_155 LRPDIPVVGSKFTIALVREKCR-EHRLKPKFVEVAERQSSQHGVFECEYF
TH_0833|M.thermoresistible__bu LRPDIPVVGSEFTLALVREKCR-EHRIKPKLVQVAEGQRSRHGVFEAQYF
MLBr_01512|M.leprae_Br4923 LRSDIPVVGSKFTLALVAAKCR-EHRIKPVFVEVVGGQSSRHGVFECEYF
MAB_3083c|M.abscessus_ATCC_199 MRPDIPVVGSAFTLALVEAKCR-EHRITPRFIRVTEGERSTHGPFECEYF
Mvan_4121|M.vanbaalenii_PYR-1 DGFRGPIVSTEATAAVADIILRDSAKLQERDAEFLNRHKATKHHPALPLY
*:*.: * *: * . :: .. : : :
Mflv_3999|M.gilvum_PYR-GCK AVNHSVPGCLAIGIHTGAGTVLHTGDIKLDQLPPDGRPTDLPGMSRLGDA
MSMEG_2685|M.smegmatis_MC2_155 AVNHSIPGCLAVAIHTGAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDA
TH_0833|M.thermoresistible__bu AVNHSIPGALAIALHTRAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDE
MLBr_01512|M.leprae_Br4923 AVNHSIPDALTIAVYTGAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDA
MAB_3083c|M.abscessus_ATCC_199 AVNHSIPDALAIAVHTEAGTILHTGDIKLDPLPPDRRPTDLPGLSGLGDR
Mvan_4121|M.vanbaalenii_PYR-1 DSDDAQRALELFDAHPFGEEFNLPGRGPAVTFRRAGHILGAATVDLAWHG
:.: . :. . . .* : : . . :. .
Mflv_3999|M.gilvum_PYR-GCK GVDLFLCDSTNSEIPGVGPSESEVGPALHRLMRGADGRVIVACFASNVDR
MSMEG_2685|M.smegmatis_MC2_155 GVDLFLCDSTNSEHPGVSPSESEVGPTLHRLIRGAEGRVIVACFASNVDR
TH_0833|M.thermoresistible__bu GVDLFLCDSTNSEIPGVGPSESEVGPNLHRLIRGAEGRVIIACFASNVDR
MLBr_01512|M.leprae_Br4923 GVDLFLCDSTNAEIPGVGPSESEVGPTLHRLIQGADGRVIVACFASNVDR
MAB_3083c|M.abscessus_ATCC_199 GVDLFLVDSTNSEVPGVGPSESEIAPTLNRLIETARGRVIVACFASNVAR
Mvan_4121|M.vanbaalenii_PYR-1 RRIVFTGDLGRYDDP--VMLDPEPVPAADYLVMESTYGDRLHGHADPAEA
:* * . : * :.* * . *: : : .*. .
Mflv_3999|M.gilvum_PYR-GCK VQSIIDAAVALGRRVSFVGRSMVRNMGIARELGYLKVADEDVLDIAAAEM
MSMEG_2685|M.smegmatis_MC2_155 VQQIIDAAVALGRRVSFVGRSMVRNMGIARELGYLKVDDSDILDIAAAEM
TH_0833|M.thermoresistible__bu VQQIIDAAVALGRRVAFVGRSMVRNMTIARELGYLRVADEDLLDIGAAEM
MLBr_01512|M.leprae_Br4923 VQQVIDAAVALGRRVSFVGRSMVRNMSIARDLGFLRVDDSHMIDIAAAEM
MAB_3083c|M.abscessus_ATCC_199 VQQIIDASVAHGRRVALVGRSMVRNMGIARELGFLEVDDRDLINVEAANE
Mvan_4121|M.vanbaalenii_PYR-1 LAEVIDATVDRGGTVVIPAFAVGRAQALLYYLWRLHSAGR----------
: .:***:* * * : . :: * : * *. .
Mflv_3999|M.gilvum_PYR-GCK MPADRVVLITTGTQGEPMAALSRMSRGEHRSITLTAGDLIILSSSLIPGN
MSMEG_2685|M.smegmatis_MC2_155 MPPDRVVLITTGTQGEPMAALSRMSRGEHRSITLTSGDLIILSSSLIPGN
TH_0833|M.thermoresistible__bu MPPERVVLITTGTQGEPMSALSRMSRGEHRSITLTPGDLVVLSSSLIPGN
MLBr_01512|M.leprae_Br4923 MAPEQVVLITTGTQGEPMSALSRMSRGEHQSITLTSGDLVVLSSSLIPGN
MAB_3083c|M.abscessus_ATCC_199 LAPQRVVLITTGTQGEPMAALSRMARGDHRQITLTPDDLVILSSSLVPGN
Mvan_4121|M.vanbaalenii_PYR-1 -LPGIPIYLDSPMAINASDLLGAFPR-DHRLPPEVYDDMCALATYTRDTD
. : : : :. *. :.* :*: . . .*: *:: :
Mflv_3999|M.gilvum_PYR-GCK EEAVYGVIDSLAKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVM
MSMEG_2685|M.smegmatis_MC2_155 EEAVYGVIDSLSKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVM
TH_0833|M.thermoresistible__bu EEAVYGVIDALAKIGARVVTNQQVRVHVSGHAYAGELLFLYNGVRPRNVM
MLBr_01512|M.leprae_Br4923 EEAVYGVIDALAKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRQVM
MAB_3083c|M.abscessus_ATCC_199 EEAVYGIIDSLAKVGTRVVTNTHARVHVSGHAYAGELLYLYNAVRPRNVM
Mvan_4121|M.vanbaalenii_PYR-1 E-----------SKKITASREPKIVISASGMATGGRILHHLKAFAPDPRN
* . . : : : .** * .*.:*. :.. *
Mflv_3999|M.gilvum_PYR-GCK PVHGTWRMLRANAALAARTGVPDENIVIAENGVSVDLVAGRVSVAGSVPV
MSMEG_2685|M.smegmatis_MC2_155 PVHGTWRHLRANAALAASTGVPPENIVLAENGVSVDLVAGRASISGAVTV
TH_0833|M.thermoresistible__bu PVHGTWRHLRANAALAVRTGVPPESVLLAENGVSVDLVAGKARIAGAVPV
MLBr_01512|M.leprae_Br4923 PVHGTWRMLRANAKLAASTGVPEDSILIAENGVSVDLVAGKASVSGAVPV
MAB_3083c|M.abscessus_ATCC_199 PVHGTWRHLRANAGLAAKTGVEEGRIVLAENGVSVDLHNERARISGRVPV
Mvan_4121|M.vanbaalenii_PYR-1 TIIVTGYQVPGTRGRSIAAGEP-----------YVKIHGEWIPINAQVAN
.: * : .. : :* *.: : . *.
Mflv_3999|M.gilvum_PYR-GCK GKMFVDGLVTGDVGDATLGERLILSSG-FVAVTVVVRRGTGKAVAPAHLH
MSMEG_2685|M.smegmatis_MC2_155 GKMFVDGLITGDVGDATLGERLILSSG-FVSITVVVHRGTGRPAGPAHLI
TH_0833|M.thermoresistible__bu GKMFVDGLITGDVGDATLGERLILSSG-FVAVTVVVRRGTGRPAGPAHLL
MLBr_01512|M.leprae_Br4923 GKMFVDGLITGDVGDVILGERLILSSG-FVAVTVVVKRSTGRPVTVPQLY
MAB_3083c|M.abscessus_ATCC_199 GKMFVHGLVDGDVSDATVGERLVLGNGGFVAVTVVVR-SNGRPVGRPQLF
Mvan_4121|M.vanbaalenii_PYR-1 LQMLS---------AHADADELIRWAGGFVTAPRQVFVVHGE--------
:*: .:.*: * **: . * *.
Mflv_3999|M.gilvum_PYR-GCK ARGFSEDPKALEPAARKVEAELEHLASQNVTEPTRIAQGVRRAVGKWVGE
MSMEG_2685|M.smegmatis_MC2_155 SRGFSEDPKALEPVAQKVERELEALAADNVTDPTRIAQAVRRTVGKWVGE
TH_0833|M.thermoresistible__bu SRGFSEDPKALEPAAQKVEEELEKLAAERVTDPGRIAQAVRRTVGKWVGE
MLBr_01512|M.leprae_Br4923 SRGFSEDPKALEPAVRKVEAELESLLAENVTDPSKIAQAVRRAIGKWVGE
MAB_3083c|M.abscessus_ATCC_199 SRGFSDDAKALDPVTELVITELNSLAEQSVNDTGRIAQSVRRVVGKWVGE
Mvan_4121|M.vanbaalenii_PYR-1 -------PQAADTLRRRLDREFG-----------------------WQAT
.:* :. . : *: * .
Mflv_3999|M.gilvum_PYR-GCK TYRRQPMIVPTVIEI-
MSMEG_2685|M.smegmatis_MC2_155 TYRRQPMIVPTVIEI-
TH_0833|M.thermoresistible__bu TYRRQPMIVPTVIEI-
MLBr_01512|M.leprae_Br4923 TYRRQPMIVPTVIEV-
MAB_3083c|M.abscessus_ATCC_199 TYRRQPMIVPTVLTVD
Mvan_4121|M.vanbaalenii_PYR-1 VPRPNQQFSL------
. * : :