For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAELAPPPPLAPGGLRVTALGGISEIGRNMTVFEHLGRLLIVDCGVLFPGHDEPGVDLILPDLRHIEDR LDEIEALVVTHAHEDHIGAIPFLLKLRPDIPVVGSKFTIALVREKCREHRLKPKFVEVAERQSSQHGVFE CEYFAVNHSIPGCLAVAIHTGAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDAGVDLFLCDSTNSEHPG VSPSESEVGPTLHRLIRGAEGRVIVACFASNVDRVQQIIDAAVALGRRVSFVGRSMVRNMGIARELGYLK VDDSDILDIAAAEMMPPDRVVLITTGTQGEPMAALSRMSRGEHRSITLTSGDLIILSSSLIPGNEEAVYG VIDSLSKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVMPVHGTWRHLRANAALAASTGVPPENI VLAENGVSVDLVAGRASISGAVTVGKMFVDGLITGDVGDATLGERLILSSGFVSITVVVHRGTGRPAGPA HLISRGFSEDPKALEPVAQKVERELEALAADNVTDPTRIAQAVRRTVGKWVGETYRRQPMIVPTVIEI
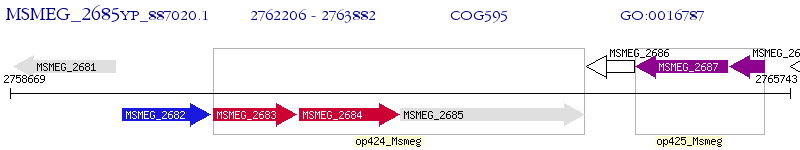
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2685 | - | - | 100% (558) | metallo-beta-lactamase superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2773c | - | 0.0 | 84.95% (558) | hypothetical protein Mb2773c |
| M. gilvum PYR-GCK | Mflv_3999 | - | 0.0 | 87.86% (552) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv2752c | - | 0.0 | 84.95% (558) | hypothetical protein Rv2752c |
| M. leprae Br4923 | MLBr_01512 | - | 0.0 | 85.56% (554) | hypothetical protein MLBr_01512 |
| M. abscessus ATCC 19977 | MAB_3083c | - | 0.0 | 75.36% (556) | hypothetical protein MAB_3083c |
| M. marinum M | MMAR_1963 | - | 0.0 | 87.10% (558) | hydrolase |
| M. avium 104 | MAV_3643 | - | 0.0 | 86.56% (558) | hydrolase of the metallo-beta-lactamase superfamily protein |
| M. thermoresistible (build 8) | TH_0833 | - | 0.0 | 87.75% (555) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2388 | - | 0.0 | 88.71% (558) | beta-lactamase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3999|M.gilvum_PYR-GCK MDLTHTPP-----GPLAPGGLRVTALGGIGEIGRNMTVFEHLGRLLIVDC
Mvan_2388|M.vanbaalenii_PYR-1 MNSELTPP-----GPLAPGGLRVTALGGINEIGRNMTVFEHLGRLLIVDC
MSMEG_2685|M.smegmatis_MC2_155 MSAELAPP-----PPLAPGGLRVTALGGISEIGRNMTVFEHLGRLLIVDC
TH_0833|M.thermoresistible__bu MSVPIPPPDLGPPGPLERGGLRVTALGGIGEIGRNMTVFEHRGRLLIIDC
Mb2773c|M.bovis_AF2122/97 MDVDLPPP-----GPLTSGGLRVTALGGINEIGRNMTVFEHLGRLLIIDC
Rv2752c|M.tuberculosis_H37Rv MDVDLPPP-----GPLTSGGLRVTALGGINEIGRNMTVFEHLGRLLIIDC
MMAR_1963|M.marinum_M MNEELPPP-----APLASGGLRVTALGGINEIGRNMTVFEHLGRLLIIDC
MLBr_01512|M.leprae_Br4923 MDVALAPP-----GPLASGGLRVTALGGISEIGRNMTIFEHLGRLLIIDC
MAV_3643|M.avium_104 MDVDLAPP-----GPLAAGGLRVTALGGISEIGRNMTVFEHLGRLLIIDC
MAB_3083c|M.abscessus_ATCC_199 MAVTPPED-LRAPGPLADGALRVTALGGISEIGRNMTVFELRGRLLIIDC
* . ** *.*********.*******:** *****:**
Mflv_3999|M.gilvum_PYR-GCK GVLFPTHDEPGVDLILPDLRHVQDRLDDVEAIVVTHAHEDHIGAIPFVLK
Mvan_2388|M.vanbaalenii_PYR-1 GVLFPTHDEPGVDLILPDLRHLENRLDDIEALVVTHGHEDHIGAIPFLLK
MSMEG_2685|M.smegmatis_MC2_155 GVLFPGHDEPGVDLILPDLRHIEDRLDEIEALVVTHAHEDHIGAIPFLLK
TH_0833|M.thermoresistible__bu GVLFPGHDEPGVDLILPDLRHIENRLDDVEALLLTHAHEDHIGAVPFLLK
Mb2773c|M.bovis_AF2122/97 GVLFPGHDEPGVDLILPDMRHVEDRLDDIEALVLTHGHEDHIGAIPFLLK
Rv2752c|M.tuberculosis_H37Rv GVLFPGHDEPGVDLILPDMRHVEDRLDDIEALVLTHGHEDHIGAIPFLLK
MMAR_1963|M.marinum_M GVLFPTHDEPGVDLILPDLRHIEDRLDDIEALVLTHAHEDHIGAIPFLLR
MLBr_01512|M.leprae_Br4923 GVLSPGHDEPGVDLILPDLRHIEDRLDDIEALVLTHAHEDHIGAIPFLLK
MAV_3643|M.avium_104 GVMFPTHDEPGVDLILPDLRHIEDRLDDIEALVLTHAHEDHIGGIPYLLK
MAB_3083c|M.abscessus_ATCC_199 GVLFPGHNEPGVDLILPDIRHIEDRLEDVEALVLTHAHEDHIGAIPFLLK
**: * *:**********:**:::**:::**:::**.******.:*::*:
Mflv_3999|M.gilvum_PYR-GCK QRADIPIVGSKFTLALIAEKCREHRLSPKLVEVTEGTKSTHGVFECQYFA
Mvan_2388|M.vanbaalenii_PYR-1 QRADIPIVGSKFTLALVAEKCREHRLSPKLVEVAEGQKSTHGVFECQYFA
MSMEG_2685|M.smegmatis_MC2_155 LRPDIPVVGSKFTIALVREKCREHRLKPKFVEVAERQSSQHGVFECEYFA
TH_0833|M.thermoresistible__bu LRPDIPVVGSEFTLALVREKCREHRIKPKLVQVAEGQRSRHGVFEAQYFA
Mb2773c|M.bovis_AF2122/97 LRPDIPVVGSKFTLALVAEKCREYRITPVFVEVREGQSTRHGVFECEYFA
Rv2752c|M.tuberculosis_H37Rv LRPDIPVVGSKFTLALVAEKCREYRITPVFVEVREGQSTRHGVFECEYFA
MMAR_1963|M.marinum_M LRPDIPIVGSKFTLALVAAKCREYRIKPVFVEVAEGQSSRHGVFECEYFA
MLBr_01512|M.leprae_Br4923 LRSDIPVVGSKFTLALVAAKCREHRIKPVFVEVVGGQSSRHGVFECEYFA
MAV_3643|M.avium_104 LRPDIPVVGSKFTLALVAAKCREHRLKPVFVEVSEGRKSTHGVFECEYFA
MAB_3083c|M.abscessus_ATCC_199 MRPDIPVVGSAFTLALVEAKCREHRITPRFIRVTEGERSTHGPFECEYFA
*.***:*** **:**: ****:*:.* ::.* : ** **.:***
Mflv_3999|M.gilvum_PYR-GCK VNHSVPGCLAIGIHTGAGTVLHTGDIKLDQLPPDGRPTDLPGMSRLGDAG
Mvan_2388|M.vanbaalenii_PYR-1 VNHSIPGCLAIGIHTGAGTVLHTGDIKLDQLPPDGRPTDLPGMSRLGDAG
MSMEG_2685|M.smegmatis_MC2_155 VNHSIPGCLAVAIHTGAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDAG
TH_0833|M.thermoresistible__bu VNHSIPGALAIALHTRAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDEG
Mb2773c|M.bovis_AF2122/97 VNHSTPDALAIAVYTGAGTILHTGDIKFDQLPPDGRPTDLPGMSRLGDTG
Rv2752c|M.tuberculosis_H37Rv VNHSTPDALAIAVYTGAGTILHTGDIKFDQLPPDGRPTDLPGMSRLGDTG
MMAR_1963|M.marinum_M VNHSVPDALAIAVYTGAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDAG
MLBr_01512|M.leprae_Br4923 VNHSIPDALTIAVYTGAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDAG
MAV_3643|M.avium_104 VNHSIPDALAIAVHTGAGTVLHTGDIKLDQLPLDGRPTDLPGMSRLGDAG
MAB_3083c|M.abscessus_ATCC_199 VNHSIPDALAIAVHTEAGTILHTGDIKLDPLPPDRRPTDLPGLSGLGDRG
**** *..*::.::* ***:*******:* ** * *******:* *** *
Mflv_3999|M.gilvum_PYR-GCK VDLFLCDSTNSEIPGVGPSESEVGPALHRLMRGADGRVIVACFASNVDRV
Mvan_2388|M.vanbaalenii_PYR-1 VDLFLCDSTNSEIPGVGPSESEVGPSLHRLMRGADGRVIVACFASNVDRV
MSMEG_2685|M.smegmatis_MC2_155 VDLFLCDSTNSEHPGVSPSESEVGPTLHRLIRGAEGRVIVACFASNVDRV
TH_0833|M.thermoresistible__bu VDLFLCDSTNSEIPGVGPSESEVGPNLHRLIRGAEGRVIIACFASNVDRV
Mb2773c|M.bovis_AF2122/97 VDLLLCDSTNAEIPGVGPSESEVGPTLHRLIRGADGRVIVACFASNVDRV
Rv2752c|M.tuberculosis_H37Rv VDLLLCDSTNAEIPGVGPSESEVGPTLHRLIRGADGRVIVACFASNVDRV
MMAR_1963|M.marinum_M VDLFLCDSTNSEIPGVGPSESEVGPTLHRLIRGADGRVIVACFASNVDRV
MLBr_01512|M.leprae_Br4923 VDLFLCDSTNAEIPGVGPSESEVGPTLHRLIQGADGRVIVACFASNVDRV
MAV_3643|M.avium_104 VDLFLCDSTNAELPGVGPSESEVGPTLHRLIRGAEGRVIVACFASNVDRV
MAB_3083c|M.abscessus_ATCC_199 VDLFLVDSTNSEVPGVGPSESEIAPTLNRLIETARGRVIVACFASNVARV
***:* ****:* ***.*****:.* *:**:. * ****:******* **
Mflv_3999|M.gilvum_PYR-GCK QSIIDAAVALGRRVSFVGRSMVRNMGIARELGYLKVADEDVLDIAAAEMM
Mvan_2388|M.vanbaalenii_PYR-1 QSIIDAAVALGRRVSFVGRSMVRNMGIARQLGYLKVADEDVLDIAAAEMM
MSMEG_2685|M.smegmatis_MC2_155 QQIIDAAVALGRRVSFVGRSMVRNMGIARELGYLKVDDSDILDIAAAEMM
TH_0833|M.thermoresistible__bu QQIIDAAVALGRRVAFVGRSMVRNMTIARELGYLRVADEDLLDIGAAEMM
Mb2773c|M.bovis_AF2122/97 QQIIDAAVALGRRVSFVGRSMVRNMRVARQLGFLRVADSDLIDIAAAETM
Rv2752c|M.tuberculosis_H37Rv QQIIDAAVALGRRVSFVGRSMVRNMRVARQLGFLRVADSDLIDIAAAETM
MMAR_1963|M.marinum_M QQIIDAAVALGRRVSFVGRSMVRNMGIARELGFLRVDDSDVIDIGAAEMM
MLBr_01512|M.leprae_Br4923 QQVIDAAVALGRRVSFVGRSMVRNMSIARDLGFLRVDDSHMIDIAAAEMM
MAV_3643|M.avium_104 QQIIDAAVALGRRVSFVGRSMVRNMGIARELGFLRVADADVIDIGAAELM
MAB_3083c|M.abscessus_ATCC_199 QQIIDASVAHGRRVALVGRSMVRNMGIARELGFLEVDDRDLINVEAANEL
*.:***:** ****::********* :**:**:*.* * .:::: **: :
Mflv_3999|M.gilvum_PYR-GCK PADRVVLITTGTQGEPMAALSRMSRGEHRSITLTAGDLIILSSSLIPGNE
Mvan_2388|M.vanbaalenii_PYR-1 PADRVVLITTGTQGEPMAALSRMSRGEHRSITLTAGDLIILSSSLIPGNE
MSMEG_2685|M.smegmatis_MC2_155 PPDRVVLITTGTQGEPMAALSRMSRGEHRSITLTSGDLIILSSSLIPGNE
TH_0833|M.thermoresistible__bu PPERVVLITTGTQGEPMSALSRMSRGEHRSITLTPGDLVVLSSSLIPGNE
Mb2773c|M.bovis_AF2122/97 APDQVVLITTGTQGEPMSALSRMSRGEHRSITLTAGDLIVLSSSLIPGNE
Rv2752c|M.tuberculosis_H37Rv APDQVVLITTGTQGEPMSALSRMSRGEHRSITLTAGDLIVLSSSLIPGNE
MMAR_1963|M.marinum_M PPEQVLLITTGTQGEPMSALSRMSRGEHRSITLTAGDLIVLSSSLIPGNE
MLBr_01512|M.leprae_Br4923 APEQVVLITTGTQGEPMSALSRMSRGEHQSITLTSGDLVVLSSSLIPGNE
MAV_3643|M.avium_104 PPERVVLITTGTQGEPMSALSRMSRGEHRSITLTAGDLVVLASSLIPGNE
MAB_3083c|M.abscessus_ATCC_199 APQRVVLITTGTQGEPMAALSRMARGDHRQITLTPDDLVILSSSLVPGNE
..::*:***********:*****:**:*:.****..**::*:***:****
Mflv_3999|M.gilvum_PYR-GCK EAVYGVIDSLAKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVMP
Mvan_2388|M.vanbaalenii_PYR-1 EAVYGVMDSLAKIGARVVTNQQARVHVSGHAYAGELLFLYNGVRPRNVMP
MSMEG_2685|M.smegmatis_MC2_155 EAVYGVIDSLSKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVMP
TH_0833|M.thermoresistible__bu EAVYGVIDALAKIGARVVTNQQVRVHVSGHAYAGELLFLYNGVRPRNVMP
Mb2773c|M.bovis_AF2122/97 EAVFGVIDALSKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVMP
Rv2752c|M.tuberculosis_H37Rv EAVFGVIDALSKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVMP
MMAR_1963|M.marinum_M EAVFGVIDSLSKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRNVMP
MLBr_01512|M.leprae_Br4923 EAVYGVIDALAKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRQVMP
MAV_3643|M.avium_104 EAVYGVIDELAKIGARVVTNAQARVHVSGHAYAGELLFLYNGVRPRHVMP
MAB_3083c|M.abscessus_ATCC_199 EAVYGIIDSLAKVGTRVVTNTHARVHVSGHAYAGELLYLYNAVRPRNVMP
***:*::* *:*:*:***** :.**************:***.****:***
Mflv_3999|M.gilvum_PYR-GCK VHGTWRMLRANAALAARTGVPDENIVIAENGVSVDLVAGRVSVAGSVPVG
Mvan_2388|M.vanbaalenii_PYR-1 VHGTWRMMRANAALAASTGVPEENIVLAENGVSVDLVAGRAGIAGAVPVG
MSMEG_2685|M.smegmatis_MC2_155 VHGTWRHLRANAALAASTGVPPENIVLAENGVSVDLVAGRASISGAVTVG
TH_0833|M.thermoresistible__bu VHGTWRHLRANAALAVRTGVPPESVLLAENGVSVDLVAGKARIAGAVPVG
Mb2773c|M.bovis_AF2122/97 VHGTWRMLRANAKLAASTGVPQESILLAENGVSVDLVAGKASISGAVPVG
Rv2752c|M.tuberculosis_H37Rv VHGTWRMLRANAKLAASTGVPQESILLAENGVSVDLVAGKASISGAVPVG
MMAR_1963|M.marinum_M VHGTWRMLRANAKLAASAGVAPESILLAENGVSVDLVGRKASIAGAVPVG
MLBr_01512|M.leprae_Br4923 VHGTWRMLRANAKLAASTGVPEDSILIAENGVSVDLVAGKASVSGAVPVG
MAV_3643|M.avium_104 VHGTWRMLRANSKLAARTGVPEESILLAENGVSVDLVAGRASISGAVPVG
MAB_3083c|M.abscessus_ATCC_199 VHGTWRHLRANAGLAAKTGVEEGRIVLAENGVSVDLHNERARISGRVPVG
****** :***: **. :** :::********* :. ::* *.**
Mflv_3999|M.gilvum_PYR-GCK KMFVDGLVTGDVGDATLGERLILSSG-FVAVTVVVRRGTGKAVAPAHLHA
Mvan_2388|M.vanbaalenii_PYR-1 KMFVDGQITGDVGDATLGERLILSSG-FIAVTVIVRRGTGKPAAPAHLHS
MSMEG_2685|M.smegmatis_MC2_155 KMFVDGLITGDVGDATLGERLILSSG-FVSITVVVHRGTGRPAGPAHLIS
TH_0833|M.thermoresistible__bu KMFVDGLITGDVGDATLGERLILSSG-FVAVTVVVRRGTGRPAGPAHLLS
Mb2773c|M.bovis_AF2122/97 KMFVDGLIAGDVGDITLGERLILSSG-FVAVTVVVRRGTGQPLAAPHLHS
Rv2752c|M.tuberculosis_H37Rv KMFVDGLIAGDVGDITLGERLILSSG-FVAVTVVVRRGTGQPLAAPHLHS
MMAR_1963|M.marinum_M KMFVDGLITGDVGEITLGERLILSSG-FIAVTVVVHRGTGRPAAVPHLHS
MLBr_01512|M.leprae_Br4923 KMFVDGLITGDVGDVILGERLILSSG-FVAVTVVVKRSTGRPVTVPQLYS
MAV_3643|M.avium_104 KMFVDGLVDGDVGDITLGERLILSSG-FVAVTVVVQRGTGRPVAPPHLHS
MAB_3083c|M.abscessus_ATCC_199 KMFVHGLVDGDVSDATVGERLVLGNGGFVAVTVVVR-SNGRPVGRPQLFS
****.* : ***.: :****:*..* *:::**:*: ..*:. .:* :
Mflv_3999|M.gilvum_PYR-GCK RGFSEDPKALEPAARKVEAELEHLASQNVTEPTRIAQGVRRAVGKWVGET
Mvan_2388|M.vanbaalenii_PYR-1 RGFSEDPKALEPVARKVGAELDQLASQNVTDPARIAQAVRRTVGKWVGET
MSMEG_2685|M.smegmatis_MC2_155 RGFSEDPKALEPVAQKVERELEALAADNVTDPTRIAQAVRRTVGKWVGET
TH_0833|M.thermoresistible__bu RGFSEDPKALEPAAQKVEEELEKLAAERVTDPGRIAQAVRRTVGKWVGET
Mb2773c|M.bovis_AF2122/97 RGFSEDPKALEPAVRKVEAELESLVAANVTDPIRIAQGVRRTVGKWVGET
Rv2752c|M.tuberculosis_H37Rv RGFSEDPKALEPAVRKVEAELESLVAANVTDPIRIAQGVRRTVGKWVGET
MMAR_1963|M.marinum_M RGFSEDPKALEPAVRKVEAELESLVAANVTDPIRIAQGVRRTVGKWVGET
MLBr_01512|M.leprae_Br4923 RGFSEDPKALEPAVRKVEAELESLLAENVTDPSKIAQAVRRAIGKWVGET
MAV_3643|M.avium_104 RGFSEDPKALEPAVRRVEEELEALVAENITDPSRIAQGVRRTVGKWVGET
MAB_3083c|M.abscessus_ATCC_199 RGFSDDAKALDPVTELVITELNSLAEQSVNDTGRIAQSVRRVVGKWVGET
****:*.***:*... * **: * :.:. :***.***.:*******
Mflv_3999|M.gilvum_PYR-GCK YRRQPMIVPTVIEI-
Mvan_2388|M.vanbaalenii_PYR-1 YRRQPMIVPTVIEI-
MSMEG_2685|M.smegmatis_MC2_155 YRRQPMIVPTVIEI-
TH_0833|M.thermoresistible__bu YRRQPMIVPTVIEI-
Mb2773c|M.bovis_AF2122/97 YRRQPMIVPTVIEV-
Rv2752c|M.tuberculosis_H37Rv YRRQPMIVPTVIEV-
MMAR_1963|M.marinum_M YRRQPMIVPTVIEV-
MLBr_01512|M.leprae_Br4923 YRRQPMIVPTVIEV-
MAV_3643|M.avium_104 YRRQPMIVPTVIEV-
MAB_3083c|M.abscessus_ATCC_199 YRRQPMIVPTVLTVD
***********: :