For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKKLVIDPVTRIEGHGKVTVHLDDDGNVVDARLHVVEFRGFEKFVQGHPYWEAPMLMQRICGICFVSHHL CGAKVLDDMIGAGTAAEVRVTRTAEKIRRLGHYAQMLQSHATAYFYLVVPEMMFGMDAAPEQRNLLGLIE ANPELMRRVIMLRKWGQEVIKTVFGRRMHGINSVPGGVDKNLSRAECDRLLDGEEGLASFDDVMDYAQDG LRLFYDFHEKHRRQVDGFAKVPALNMCLVDADGNVDYYHGALRIVDENKCIVREFDYHDYLDHFSEAVEP WSYMKFPFLKDLGREEGSVRVGPLARMNVTNSLSTPLAQEALERFHAYTGGSPNNMTLHTNWARAIEVLH AAELIGELLRDPELHGDDLVVTPGGDAWVGEGIGVVEAPRGTLLHHYRAGVGGDITFANLIVATTHNNQV LNRTVRSVAEDYLVGHGEITEGMMNAIEVGIRAFDPCLSCATHALGQMPLIVTVHGPAGDVVDERVRS*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
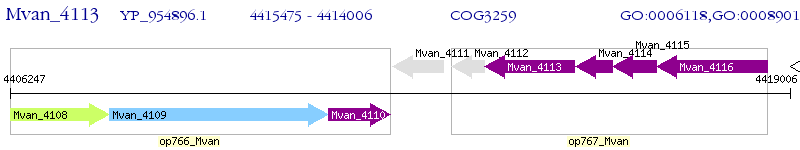
| M. vanbaalenii PYR-1 | Mvan_4113 | - | - | 100% (489) | nickel-dependent hydrogenase, large subunit |
| M. vanbaalenii PYR-1 | Mvan_2366 | - | 8e-08 | 29.57% (186) | hydrogen:quinone oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3497 | - | 4e-31 | 28.09% (477) | coenzyme F420-reducing hydrogenase, alpha subunit |
| M. avium 104 | MAV_2682 | - | 2e-07 | 26.02% (196) | hydrogen:quinone oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_3928 | - | 2e-31 | 28.51% (477) | [NiFe] hydrogenase, alpha subunit, putative [Mycobacterium |
| M. thermoresistible (build 8) | TH_1153 | - | 6e-31 | 28.48% (467) | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3497|M.marinum_M -------------MAPDNRTLSVGTLTRVEGEGALYVSLKD-GELDRVEL
MSMEG_3928|M.smegmatis_MC2_155 -------------MNPEVRTLSVGALARVEGEGALHVTLRD-GAVVGTQL
TH_1153|M.thermoresistible__bu -------------VTPEIRTLTVGALTRVEGEGALRITLRD-GQVHRAEL
Mvan_4113|M.vanbaalenii_PYR-1 ----------------MSKKLVIDPVTRIEGHGKVTVHLDDDGNVVDARL
MAV_2682|M.avium_104 MTTIIPKPSHAKRGPGQLVEMAWDPITRIVGSLGIYTKIDFDNREVVECH
: ..::*: * : : .
MMAR_3497|M.marinum_M NIYEPPRFFEAFLRGRAHTEPPDLTARICGICPVAYQVSACNALEGACG-
MSMEG_3928|M.smegmatis_MC2_155 NIYEPPRFFEAFLRGRAHTEPPDLTARVCGICPVAYQVSACNAIEDACG-
TH_1153|M.thermoresistible__bu NIYEPPRFFEAFLRGRAHTEPPDLTARVCGICPVAYQVSACNALEDACG-
Mvan_4113|M.vanbaalenii_PYR-1 HVVEF-RGFEKFVQGHPYWEAPMLMQRICGICFVSHHLCGAKVLDDMIGA
MAV_2682|M.avium_104 STSSIFRGYSIFMKGKDPRDAHFITSRICGICGDNHCTCSVYTQNMAYG-
. * :. *::*: :. : *:**** : .. . : *
MMAR_3497|M.marinum_M -----VELDADLIELRRLLYCGEWIHSHVLHICLLHAPDFLG--------
MSMEG_3928|M.smegmatis_MC2_155 -----VTVDPEIVQLRRLLYCGEWIHSHALHIFLLHLPDFLG--------
TH_1153|M.thermoresistible__bu -----VRLEPDLLALRRLLYCGEWISSHALHIVMLHAPDFLG--------
Mvan_4113|M.vanbaalenii_PYR-1 GTAAEVRVTRTAEKIRRLGHYAQMLQSHATAYFYLVVPEMMFGMDAAP--
MAV_2682|M.avium_104 -----VQPPHLAEWITNLGEAAEYMFDHNIFQENLVGVDYCEKMVKETNP
* : .* .: : .* * :
MMAR_3497|M.marinum_M ------------------YPDAIAFARDE----REIVERGLRLKKAGNQL
MSMEG_3928|M.smegmatis_MC2_155 ------------------HPDGISLAREH----PELVERGLSLKKTGNRI
TH_1153|M.thermoresistible__bu ------------------YPDAIVMAGDH----PRFVGRGMALKKAGNRL
Mvan_4113|M.vanbaalenii_PYR-1 -----------------EQRNLLGLIEAN----PELMRRVIMLRKWGQEV
MAV_2682|M.avium_104 GVLAKAENTAAPHAGAHGYKTIADIMRSLNPFSGEFYREALQVSRMTREM
: .: . : : : ..:
MMAR_3497|M.marinum_M MEFVGGRAIHPINVRLGGFYSVPTRSDFAPMAEQLR--------------
MSMEG_3928|M.smegmatis_MC2_155 MEQIGGRAIHPVNVRLGGFYSAPKPGDLKSLAALLR--------------
TH_1153|M.thermoresistible__bu LEQIGGRAVHPINMRLGGFYSVPTRAELGPLAETLR--------------
Mvan_4113|M.vanbaalenii_PYR-1 IKTVFGRRMHGINSVPGGVDKNLSRAECDRLLDGEEGLASFD--------
MAV_2682|M.avium_104 FCLMEGRHVHPSTLYPGGVGTVATIQLMTDYMSRLMRYVEFMKKVVPMHD
: : ** :* . **. . .
MMAR_3497|M.marinum_M --------------------------------------------------
MSMEG_3928|M.smegmatis_MC2_155 --------------------------------------------------
TH_1153|M.thermoresistible__bu --------------------------------------------------
Mvan_4113|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2682|M.avium_104 DLFDFFYEALPGYEKSGLRRTLLACWGSYQDPEHCNFSYKDMEKWGRKMF
MMAR_3497|M.marinum_M -------------RALDDALATVEWVAGFEFPDLEFDHEFLALTTPDRYP
MSMEG_3928|M.smegmatis_MC2_155 -------------KSLDDALYTVREVADLDCPDVVFDHEFLALGTADAYP
TH_1153|M.thermoresistible__bu -------------RALDDASYIARAVAGFDFPDVEFDHELLALGAPDRYP
Mvan_4113|M.vanbaalenii_PYR-1 -------------DVMDYAQDGLRLFYDFHEKHRRQVDGFAKVPALNMCL
MAV_2682|M.avium_104 VTPGIVVDGKLVTTSLVDINLGIRILLGSSYYDDWTDQEMFVKTDPLGNP
: . . . . . :
MMAR_3497|M.marinum_M IENG-------------TIARSAGPVFPVDEFTDHVVELQVP--------
MSMEG_3928|M.smegmatis_MC2_155 IESG-------------AIVRSAGPSFPLADFTTHVGEAQVP--------
TH_1153|M.thermoresistible__bu IEDG-------------VIRRTTGEPFGPSEFADHVVEAQVP--------
Mvan_4113|M.vanbaalenii_PYR-1 VDADGNVDYYHGALRIVDENKCIVREFDYHDYLDHFSEAVEP--------
MAV_2682|M.avium_104 VDRR------------HPWNQHTNPHPQKRDMEDKYSWVMSPRWFDGKDH
:: : : : *
MMAR_3497|M.marinum_M ---------HSTALHATLD-----GGCHVTGPLARYALNSSLLT-PIAAQ
MSMEG_3928|M.smegmatis_MC2_155 ---------HSTALHAALD-----DGRYLTGPLARYSLNSAHLP-PIARE
TH_1153|M.thermoresistible__bu ---------HSTALHATLD-----GRRYITGPLARYNLNYAQLS-PLARQ
Mvan_4113|M.vanbaalenii_PYR-1 ---------WSYMKFPFLKDLGREEGSVRVGPLARMNVTNSLST-PLAQE
MAV_2682|M.avium_104 LALDTGGGPLARLWSTALAGLVDIGYVKATGRSVQINLPKTALKGPVELE
: . * .* .: : : *: :
MMAR_3497|M.marinum_M AAARAGLSAQCRN---PFRSIIVRAVEVVYAIEEALRIIENYQRPARPFV
MSMEG_3928|M.smegmatis_MC2_155 AATSAGLGAQCRN---PFRSIVVRAVEVVFAIEEALRIIAEYQEPPRPFV
TH_1153|M.thermoresistible__bu LAAEAGLGARCWN---PFRSIVVRAVEVVYAIEEALRIIDGYRRPDRPYV
Mvan_4113|M.vanbaalenii_PYR-1 ALERFHAYTGGSPNNMTLHTNWARAIEVLHAAELIGELLRDPELHGDDLV
MAV_2682|M.avium_104 WKIPEHGSNTIERNRARTYFQAYSAAAALHFAEKALVEIRAGRTKTWEKF
* .:. * : . .
MMAR_3497|M.marinum_M EVP---ARAGVGHGVSEAPRGLLYHRYRIDDAGLVSAATIIPPTSQNQAA
MSMEG_3928|M.smegmatis_MC2_155 EVP---ARAGVGHGVSEAPRGLLYHRYEIGHDGLVRTATLIPPTAQNQAA
TH_1153|M.thermoresistible__bu EVP---ARPGVGFGVSEAPRGLLYHRYEIGADGLIAAADIVPPTSQNQAA
Mvan_4113|M.vanbaalenii_PYR-1 VTPGGDAWVGEGIGVVEAPRGTLLHHYRAGVGGDITFANLIVATTHNNQV
MAV_2682|M.avium_104 EVP----DEGIGCGFTEAVRGVLSHHMVIRDGKIANYHPYPPTPWNANPR
.* * * *. ** ** * *: .. : :
MMAR_3497|M.marinum_M IEADLARVVSENLALD---DEVLTT-----MCERAIRNYDPCISCSTHFL
MSMEG_3928|M.smegmatis_MC2_155 IEHELAVLVAANLDRD---DAELTQ-----LCERAIRNHDPCISCATHFL
TH_1153|M.thermoresistible__bu IEHEMKRLVESDPTLG---DAELTA-----LCERSIRNWDPCISCSTHFL
Mvan_4113|M.vanbaalenii_PYR-1 LNRTVRSVAEDYLVGHGEITEGMMN-----AIEVGIRAFDPCLSCATHAL
MAV_2682|M.avium_104 DVYGTPGPYEDAVQGQPIFEENDRDNFKGIDIMRTVRSFDPCLPCGVHMY
:* ***:.*..*
MMAR_3497|M.marinum_M --------TLTLERG---------
MSMEG_3928|M.smegmatis_MC2_155 --------TLTLDRG---------
TH_1153|M.thermoresistible__bu --------TLEVDRGDR-------
Mvan_4113|M.vanbaalenii_PYR-1 G-QMPLIVTVHGPAGDVVDERVRS
MAV_2682|M.avium_104 LGKGKTLDRLHSPTQTATGE----
: