For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPLDPTVVGTELPTATMTVDRGRLRFFAKAIGETDPKYTDVAAARAAGHPDLPVPPTFLFSIELEAPDPF SYLSALGVDLRRVLHGEQSFVYHAMAYPGDELLATPRISDVYTKKAGALDFIAKATRITRADGTVIADLQ SLIVIQNPGGTA
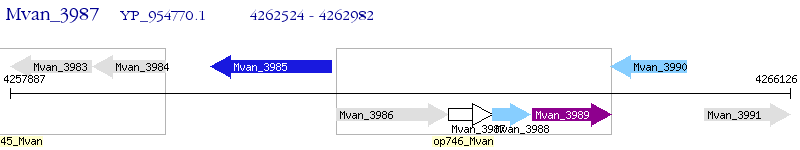
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3987 | - | - | 100% (152) | hypothetical protein Mvan_3987 |
| M. vanbaalenii PYR-1 | Mvan_5844 | - | 3e-07 | 26.57% (143) | dehydratase |
| M. vanbaalenii PYR-1 | Mvan_1232 | - | 2e-06 | 25.83% (120) | (3R)-hydroxyacyl-ACP dehydratase subunit HadC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0656 | - | 3e-06 | 26.12% (134) | (3R)-hydroxyacyl-ACP dehydratase subunit HadC |
| M. gilvum PYR-GCK | Mflv_0994 | - | 2e-08 | 27.97% (143) | dehydratase |
| M. tuberculosis H37Rv | Rv0637 | - | 3e-06 | 26.12% (134) | (3R)-hydroxyacyl-ACP dehydratase subunit HadC |
| M. leprae Br4923 | MLBr_01908 | - | 6e-06 | 26.56% (128) | (3R)-hydroxyacyl-ACP dehydratase subunit HadC |
| M. abscessus ATCC 19977 | MAB_2054 | - | 4e-22 | 39.07% (151) | hypothetical protein MAB_2054 |
| M. marinum M | MMAR_0832 | - | 6e-08 | 29.13% (127) | hypothetical protein MMAR_0832 |
| M. avium 104 | MAV_1945 | - | 2e-11 | 31.06% (132) | hypothetical protein MAV_1945 |
| M. smegmatis MC2 155 | MSMEG_1342 | - | 2e-10 | 29.37% (126) | (3R)-hydroxyacyl-ACP dehydratase subunit HadC |
| M. thermoresistible (build 8) | TH_0193 | - | 3e-07 | 27.91% (129) | MaoC like domain protein |
| M. ulcerans Agy99 | MUL_4576 | - | 4e-08 | 29.13% (127) | hypothetical protein MUL_4576 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3987|M.vanbaalenii_PYR-1 -----MP--LDPTVVGTELPTATMTVDRGRLRFFAKAIGETDPKYTDVAA
MAB_2054|M.abscessus_ATCC_1997 -----MTRVADASLIGTQLGSTTFPVDRSKVREFALSLDDHDPIYQDAAA
MAV_1945|M.avium_104 -----MTGSPAPDVDLDSGQPWEVVVERGKIAEFAEAMLSDDPAYRGPGA
Mb0656|M.bovis_AF2122/97 ---MALKTDIRGMI---WRYPDYFIVGREQCREFARAVKCDHPAFFSEEA
Rv0637|M.tuberculosis_H37Rv ---MALKTDIRGMI---WRYPDYFIVGREQCREFARAVKCDHPAFFSEEA
MLBr_01908|M.leprae_Br4923 ---MALKTDIRGMV---WRYSDYFIVGREQCREFARAIKCDHPAYFSEDA
MSMEG_1342|M.smegmatis_MC2_155 ---MALKTDIKGMV---WKYPDPFLIGREQIRQYAKAVKAMDPASHDEAA
MMAR_0832|M.marinum_M ---MTVPPEAAGLIGKHYQAPDYFVVGREKIREFATAIQDEHPSHSDEAA
MUL_4576|M.ulcerans_Agy99 ---MTVPPEAAGLIGKHYQAPDYFVVGREKIREFATAIQDEHPSHSDEAA
Mflv_0994|M.gilvum_PYR-GCK MTAPADSSPLEARVGHHYQMAGTYLVGREKVREYARAVQDYHPAHWDVAA
TH_0193|M.thermoresistible__bu MTT-AESPALEARVGHWYQMDNTYLVGREKLREYARAVQDYHPAHWDVAA
: : * : :* :: .* . *
Mvan_3987|M.vanbaalenii_PYR-1 ARAAGHPDLPVPPTFLFS-IELEAPDPFSYLSALGVDLRRVLHGEQSFVY
MAB_2054|M.abscessus_ATCC_1997 ARAAGFGAIPAPPTFVVSSAHWRADD--DMFGALGLDLRRVLHGECGWEY
MAV_1945|M.avium_104 I---------IPPTFLTSAARWAPPG---VRVSVGFDRKRLLHGEQEYTF
Mb0656|M.bovis_AF2122/97 AADLGYDALVAPLTFVTILAKYVQLD-FFRHVDVGMETMQIVQVDQRFVF
Rv0637|M.tuberculosis_H37Rv AADLGYDALVAPLTFVTILAKYVQLD-FFRHVDVGMETMQIVQVDQRFVF
MLBr_01908|M.leprae_Br4923 AAELGYDAIVAPLTFVTIFAKYVQLD-FFRNVDVGMETMQIVQVDQRFVF
MSMEG_1342|M.smegmatis_MC2_155 AAELGHPALVAPLTFASTLALLVQQH-FFMHVDVGMQTMQIVQVDQKFIY
MMAR_0832|M.marinum_M AAELGYPELVAPLTFLAIAGRRVQLE-IFNKFNIPINIARVFHRDQKFRF
MUL_4576|M.ulcerans_Agy99 ATELGYPELVAPLTFLAIAGRRVQLE-IFNKFNIPINIARVFHRDQKFRF
Mflv_0994|M.gilvum_PYR-GCK AQALGFPDVVAPLTFTSAPGMQCNRR-MFEEIVVGYDTY--LQTEEVFEQ
TH_0193|M.thermoresistible__bu AAELGYSDLVAPLTFTSAPGMTCNRE-MFESVVVGYDTY--MQTEEVFEQ
* ** : : .: : :
Mvan_3987|M.vanbaalenii_PYR-1 H-AMAYPGDELLATPRISDVYT---KKAGALDFIAKATRITRADGTVIAD
MAB_2054|M.abscessus_ATCC_1997 F-APVVVGDELTETRRVSNVTSREGKRGGTMTIVTIETDFTNQRDELVVR
MAV_1945|M.avium_104 HGDLPAEGDVLTAVERVVDRFSKPGKRGGTMRFATVVTEYRNAAGELIAE
Mb0656|M.bovis_AF2122/97 H-KPVLAGDKLWARMDIHSVDER-----FGADIVVTRNLCTNDDGELVME
Rv0637|M.tuberculosis_H37Rv H-KPVLAGDKLWARMDIHSVDER-----FGADIVVTRNLCTNDDGELVME
MLBr_01908|M.leprae_Br4923 H-KPVLVGDKLWARMDIHSVSER-----FGADIVVTKNSCTSDDGELVME
MSMEG_1342|M.smegmatis_MC2_155 H-KPLLAGDQLHAVMEIMSHEER-----FGADIVVTRNICTNDDGEVVLE
MMAR_0832|M.marinum_M Y-RPIVTGDKLYFHTYLDSVLES-----HGTVIAEIRSEVTDDEGKPVVT
MUL_4576|M.ulcerans_Agy99 Y-RPIVTGDKLYFHTYLDSVLES-----HGTVIAEIRSEVTDDEGKPVVT
Mflv_0994|M.gilvum_PYR-GCK H-RPIVAGDELVIDVELTSVRRT-----AGRDFITVTNTFTDPAGERVHT
TH_0193|M.thermoresistible__bu H-RPIVAGDELTIDVELTSVRRI-----AGRDLITVTNTFTDTAGEVVHT
. ** * : . : . . :
Mvan_3987|M.vanbaalenii_PYR-1 LQSLIVIQNPGGTA------------------------------------
MAB_2054|M.abscessus_ATCC_1997 -QTDVLIETGGSTQ------------------------------------
MAV_1945|M.avium_104 AKATFIETAAKR--------------------------------------
Mb0656|M.bovis_AF2122/97 AYTTLMGQQGDGSARLKWDKESGQVIRTA---------------------
Rv0637|M.tuberculosis_H37Rv AYTTLMGQQGDGSARLKWDKESGQVIRTA---------------------
MLBr_01908|M.leprae_Br4923 AYTTLMGQQGDNSSQLKWDKESGQVIRSA---------------------
MSMEG_1342|M.smegmatis_MC2_155 AYTTLMGHEGDNSISVKWDPETGQVIRKAIGE------------------
MMAR_0832|M.marinum_M SVVTMLGEAAHHEAEADAAVAAIASI------------------------
MUL_4576|M.ulcerans_Agy99 SVVTMLGEAAHHEAEADAAVAAIASI------------------------
Mflv_0994|M.gilvum_PYR-GCK LHTTVVGVTADDIDAGVKIAVQNAMMHDMNILDIPGTDADYEKQERPGGE
TH_0193|M.thermoresistible__bu LHTTVVGVTAEDVDPAIKTAVQNAMMHDVDIFSVG--ETEYRKTVRPEGE
.:
Mvan_3987|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2054|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_1945|M.avium_104 --------------------------------------------------
Mb0656|M.bovis_AF2122/97 --------------------------------------------------
Rv0637|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01908|M.leprae_Br4923 --------------------------------------------------
MSMEG_1342|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0832|M.marinum_M --------------------------------------------------
MUL_4576|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0994|M.gilvum_PYR-GCK VRISEGGLTRTPGSVSFDGLKVGDEIPVHHTRLSRGDLVNYAGVAGDANP
TH_0193|M.thermoresistible__bu VRISDGGLTRTPGTPAFEDVQVGDELGVHHAKLSRGDLVNYAGVAGDANP
Mvan_3987|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2054|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_1945|M.avium_104 --------------------------------------------------
Mb0656|M.bovis_AF2122/97 --------------------------------------------------
Rv0637|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01908|M.leprae_Br4923 --------------------------------------------------
MSMEG_1342|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0832|M.marinum_M --------------------------------------------------
MUL_4576|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0994|M.gilvum_PYR-GCK IHWDEEIAKLAGLPDVIAHGMLTMGLGAGFVSAWSGDPGAVTRYSIRLSQ
TH_0193|M.thermoresistible__bu IHWDEDIAKLAGLPDVIAHGMLTMGLGAGFVSRWSGDPGAVTRYAVRLSA
Mvan_3987|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2054|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_1945|M.avium_104 --------------------------------------------------
Mb0656|M.bovis_AF2122/97 --------------------------------------------------
Rv0637|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01908|M.leprae_Br4923 --------------------------------------------------
MSMEG_1342|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_0832|M.marinum_M --------------------------------------------------
MUL_4576|M.ulcerans_Agy99 --------------------------------------------------
Mflv_0994|M.gilvum_PYR-GCK PAIVSAKEGADIEYSGRIKSLDPETRSGVVLVGAKSAGRKIFGLASLHIR
TH_0193|M.thermoresistible__bu PAIVSAKEGCDIEFSGRVKSLDPQTRSGVIVVTAKSGGKKIFGLATMNIR
Mvan_3987|M.vanbaalenii_PYR-1 --
MAB_2054|M.abscessus_ATCC_1997 --
MAV_1945|M.avium_104 --
Mb0656|M.bovis_AF2122/97 --
Rv0637|M.tuberculosis_H37Rv --
MLBr_01908|M.leprae_Br4923 --
MSMEG_1342|M.smegmatis_MC2_155 --
MMAR_0832|M.marinum_M --
MUL_4576|M.ulcerans_Agy99 --
Mflv_0994|M.gilvum_PYR-GCK FR
TH_0193|M.thermoresistible__bu FR