For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVSNVVRIVPAAMLVGAVVAGTPAAQADNRRLNESVAVNVYTIQQNSGCTTDLKINPQLQLAAQWHTND VLHNRALGGDIGSDGSTAQDRANKAGFVGTVAETVAINPALAISGVEIMNQWYYRPDYMAIMSNCANTQI GVWSENSLDRSVVVAVYGQPG
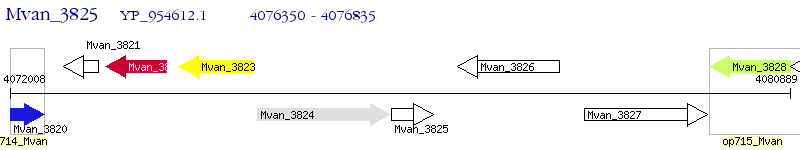
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3825 | - | - | 100% (161) | hypothetical protein Mvan_3825 |
| M. vanbaalenii PYR-1 | Mvan_0567 | - | 4e-52 | 65.22% (138) | hypothetical protein Mvan_0567 |
| M. vanbaalenii PYR-1 | Mvan_0917 | - | 5e-49 | 62.99% (154) | hypothetical protein Mvan_0917 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2711 | - | 2e-73 | 82.39% (159) | hypothetical protein Mflv_2711 |
| M. tuberculosis H37Rv | Rv1975 | - | 5e-57 | 70.92% (141) | hypothetical protein Rv1975 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2890 | - | 2e-58 | 72.11% (147) | hypothetical protein MMAR_2890 |
| M. avium 104 | MAV_1866 | - | 1e-54 | 66.45% (155) | hypothetical protein MAV_1866 |
| M. smegmatis MC2 155 | MSMEG_0355 | - | 6e-51 | 60.39% (154) | hypothetical protein MSMEG_0355 |
| M. thermoresistible (build 8) | TH_0171 | - | 4e-57 | 64.67% (150) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3914 | - | 8e-58 | 71.23% (146) | hypothetical protein MUL_3914 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1975|M.tuberculosis_H37Rv MS----RRASATCALS--ATTAVAIMAAPAARADDKRLNDGVVANVYTVQ
MMAR_2890|M.marinum_M MR----HRYLILSALV--AMVSGAALGMPAARADNKRLNDGLVANVYTVQ
MUL_3914|M.ulcerans_Agy99 MAGGHLRKQMPLPVMIGLALLSAALSTAPAAHADNKRLNDGVVANVYTIQ
Mvan_3825|M.vanbaalenii_PYR-1 -M----TVSNVVRIVPAAMLVGAVVAGTPAAQADNRRLNESVAVNVYTIQ
Mflv_2711|M.gilvum_PYR-GCK MR----PMSMVVRALPVVALVAAAVAGVPAAQADNRRLNESVVVNVYTIQ
TH_0171|M.thermoresistible__bu MT----TILWRTTATVAVLVVCQTLIGPPAA-ADNKRLNDGVVANVYTVK
MAV_1866|M.avium_104 ---------MVVMLFLVVSLTAGIA--VPSANADNKRLNESVFVNIYTAQ
MSMEG_0355|M.smegmatis_MC2_155 MN---LIAQRSVVLVACGVLTAGAVSHAPDAAADNRRLNSSVVSNVYTVQ
* * **::***..: *:** :
Rv1975|M.tuberculosis_H37Rv RQAGCTNDVTINPQLQLAAQWHTLDLLNNRHLNDDTGSDGSTPQDRAHAA
MMAR_2890|M.marinum_M HQAGCTNDVRINPQLQLAAQWHTLDVLNNRDLDGDIGSDGSAPSDRASAA
MUL_3914|M.ulcerans_Agy99 HQAGCTSELRIDPRLQLAAEWHTNDVLGNRALDGDIGSDGSTPSDRAAAA
Mvan_3825|M.vanbaalenii_PYR-1 QNSGCTTDLKINPQLQLAAQWHTNDVLHNRALGGDIGSDGSTAQDRANKA
Mflv_2711|M.gilvum_PYR-GCK HKHGCQTEIKINPQLQLAAQWHANDVLRNRALNGDIGSDGSTPQVRANNA
TH_0171|M.thermoresistible__bu KQNGCDTDIQINPQLQLAAQWHANDVLHNRTLDGDLGSDGSTVQDRADRA
MAV_1866|M.avium_104 AQNGCPGEPHLDGRLVEAARVHTLDVLHNRTLGGDIGSDGSTPQDRANRA
MSMEG_0355|M.smegmatis_MC2_155 RQAGCTDNITVDPRLLEAARRHTLDVLNNRSLDGDIGSDGSTPQVRASNA
: ** : :: :* **. *: *:* ** *..* *****: . ** *
Rv1975|M.tuberculosis_H37Rv GFRGKVAETVAINPAVAISGIELINQWYYNPAFFAIMSDCANTQIGVWSE
MMAR_2890|M.marinum_M GFHGQVAETVAINPALAISGVELLNQWYYNPAYFAIMSDCANTEIGVWSE
MUL_3914|M.ulcerans_Agy99 GFRGDVAETVAINPALAISGVELINQWYNNPDYLAIMRDCKYTAIGVWSE
Mvan_3825|M.vanbaalenii_PYR-1 GFVGTVAETVAINPALAISGVEIMNQWYYRPDYMAIMSNCANTQIGVWSE
Mflv_2711|M.gilvum_PYR-GCK GFVGTASETVAINPALAISGIEILNQWYYRPDYMAIMSNCANTEIGVWSE
TH_0171|M.thermoresistible__bu GYRGVASQTVAINPALAMSNLEVIRQWYFRPDFHAIMADCENTEIGVWSE
MAV_1866|M.avium_104 GFVGKASETVAINPALAISGIEILNQWWHNPAYQAIMRDCSNTAIGVWSE
MSMEG_0355|M.smegmatis_MC2_155 DFYGRVAETVAVNPALAINNLDVINQWYHRPDYLAIMRDCGFTRMGVWSE
.: * .::***:***:*:..::::.**: .* : *** :* * :*****
Rv1975|M.tuberculosis_H37Rv NSPDRTVVVAVYGQPDRPSAMPPRGAVTGPPSPVAAQENVPIDPSPDYDA
MMAR_2890|M.marinum_M NSPDRTVVVAMYGQLDRPVATPPVTAPTEPPPPVALPENVPIDPSPDYDA
MUL_3914|M.ulcerans_Agy99 NSLDRTVVVAVYGSPT----------------------------------
Mvan_3825|M.vanbaalenii_PYR-1 NSLDRSVVVAVYGQPG----------------------------------
Mflv_2711|M.gilvum_PYR-GCK NSLDRSVVVAVYGQPG----------------------------------
TH_0171|M.thermoresistible__bu HALDRTVVVAVYGKPGWSDR------------------------------
MAV_1866|M.avium_104 NSLDRSVVVAVYGQPG----------------------------------
MSMEG_0355|M.smegmatis_MC2_155 NSLDRSVLVAVYGSPEV---------------------------------
:: **:*:**:**.
Rv1975|M.tuberculosis_H37Rv SDEIEYGINWLPWILRGVYPPPAMPPQ
MMAR_2890|M.marinum_M SDEIEYGINWLPWILRGVYPPPAMPPE
MUL_3914|M.ulcerans_Agy99 ---------------------------
Mvan_3825|M.vanbaalenii_PYR-1 ---------------------------
Mflv_2711|M.gilvum_PYR-GCK ---------------------------
TH_0171|M.thermoresistible__bu ---------------------------
MAV_1866|M.avium_104 ---------------------------
MSMEG_0355|M.smegmatis_MC2_155 ---------------------------