For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VCNSTREKRPRGDQLDARRPNLGATKTVREASGRRLRHFLTLTVGIVAVLGVQALLITPAAHGDNKRLND GVVANVYTIQRQAGCVSDIKTSPSLQLAAQWHARDALTNRNVNGDVGSDGSTPQQRAEAAGFSGAVSETV AINPALAFSGIELINRWYHDPVDFAVMSDCSNTRIGVWSENSVDRTVVVAVYGKPQ
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
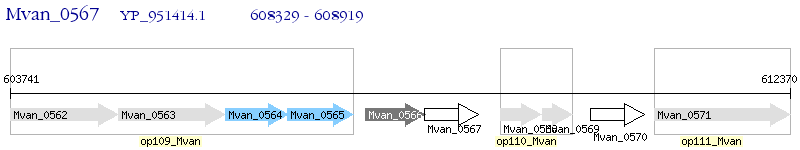
| M. vanbaalenii PYR-1 | Mvan_0567 | - | - | 100% (196) | hypothetical protein Mvan_0567 |
| M. vanbaalenii PYR-1 | Mvan_3825 | - | 6e-52 | 65.22% (138) | hypothetical protein Mvan_3825 |
| M. vanbaalenii PYR-1 | Mvan_0917 | - | 4e-47 | 56.77% (155) | hypothetical protein Mvan_0917 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0523 | - | 1e-101 | 93.91% (197) | hypothetical protein Mflv_0523 |
| M. tuberculosis H37Rv | Rv1975 | - | 1e-58 | 71.83% (142) | hypothetical protein Rv1975 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4048 | - | 4e-57 | 65.24% (164) | hypothetical protein MMAR_4048 |
| M. avium 104 | MAV_2072 | - | 3e-53 | 63.89% (144) | hypothetical protein MAV_2072 |
| M. smegmatis MC2 155 | MSMEG_0355 | - | 1e-45 | 57.43% (148) | hypothetical protein MSMEG_0355 |
| M. thermoresistible (build 8) | TH_0483 | - | 2e-57 | 63.64% (154) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3914 | - | 2e-57 | 65.24% (164) | hypothetical protein MUL_3914 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1975|M.tuberculosis_H37Rv ----------------------------------MSR--RASATCALS-A
MAV_2072|M.avium_104 ----------------------------------MSVPVIRAARLMAG-A
TH_0483|M.thermoresistible__bu ----------------------------------MNS---RITSGLAA-A
MSMEG_0355|M.smegmatis_MC2_155 ----------------------------------MNLIAQRSVVLVACGV
MMAR_4048|M.marinum_M -------------------------------MAGGHLRKQMPLPVMIGLA
MUL_3914|M.ulcerans_Agy99 -------------------------------MAGGHLRKQMPLPVMIGLA
Mvan_0567|M.vanbaalenii_PYR-1 MCNSTREKRPRGDQLDARRPNLGATKTVREASG-RRLRHFLTLTVGIVAV
Mflv_0523|M.gilvum_PYR-GCK MCNSTHEKRPRGDQLYARRPNPGAMKMVREPSGGRRLRHPLTLTVGIVAV
.
Rv1975|M.tuberculosis_H37Rv TTAVAIMAAPAARADDKRLNDGVVANVYTVQRQAGCTNDVTINPQLQLAA
MAV_2072|M.avium_104 VVVSAALPVPAAHADNKRLNSAVVSAVYTLQHQAGCTNDVVRNNALTLAA
TH_0483|M.thermoresistible__bu SIAVATMTAPTAAADNTRLNDGLVANVYTIQQQAGCETEVKKNPQLQLAA
MSMEG_0355|M.smegmatis_MC2_155 LTAGAVSHAPDAAADNRRLNSSVVSNVYTVQRQAGCTDNITVDPRLLEAA
MMAR_4048|M.marinum_M LLSAALSTAPAAHADNKRLNDGVVANVYTIQHQAGCTSELRIDPRLQLAA
MUL_3914|M.ulcerans_Agy99 LLSAALSTAPAAHADNKRLNDGVVANVYTIQHQAGCTSELRIDPRLQLAA
Mvan_0567|M.vanbaalenii_PYR-1 LGVQALLITPAAHGDNKRLNDGVVANVYTIQRQAGCVSDIKTSPSLQLAA
Mflv_0523|M.gilvum_PYR-GCK LGVQALLITPAAHGDNKRLNDGVVANVYTIQRQAGCVSDIKISPSLQLAA
* .* * .*: ***..:*: ***:*:**** :: . * **
Rv1975|M.tuberculosis_H37Rv QWHTLDLLNNRHLNDDTGSDGSTPQDRAHAAGFRGKVAETVAINPAVAIS
MAV_2072|M.avium_104 EWHAEDMINNRNINDDIGSDGSTPQDRANAAGFRGRAAETVAINPAMAIS
TH_0483|M.thermoresistible__bu EWHTRDLMNNRALDGDIGSDGSTPQQRAEAAGYRGRVAETIVIHPALAIS
MSMEG_0355|M.smegmatis_MC2_155 RRHTLDVLNNRSLDGDIGSDGSTPQVRASNADFYGRVAETVAVNPALAIN
MMAR_4048|M.marinum_M EWHTNDVLGNRALDGDIGSDGSTPSDRAAAAGFRGDVAETVAINPALAIS
MUL_3914|M.ulcerans_Agy99 EWHTNDVLGNRALDGDIGSDGSTPSDRAAAAGFRGDVAETVAINPALAIS
Mvan_0567|M.vanbaalenii_PYR-1 QWHARDALTNRNVNGDVGSDGSTPQQRAEAAGFSGAVSETVAINPALAFS
Mflv_0523|M.gilvum_PYR-GCK QWHARDALTNRNVNGDVGSDGSTLQQRAEAAGFSGAVSETVAINPALAFS
. *: * : ** ::.* ****** . ** *.: * .:**:.::**:*:.
Rv1975|M.tuberculosis_H37Rv GIELINQWYYNPAFFAIMSDCANTQIGVWSENSPDRTVVVAVYGQPDRPS
MAV_2072|M.avium_104 SLELVNQWYANPDDMAIIRDCANTSIGVWSDNSLDRTVVVAVYGQPD---
TH_0483|M.thermoresistible__bu GLELLNAWYHNPAYYAIMSDCAHTDIGVWSENSLDRTVVVAVYGQPLP--
MSMEG_0355|M.smegmatis_MC2_155 NLDVINQWYHRPDYLAIMRDCGFTRMGVWSENSLDRSVLVAVYGSPEV--
MMAR_4048|M.marinum_M GVELINQWYNNPDYLAIMRDCKYTAIGVWSENSLDRTVVVAVYGSPT---
MUL_3914|M.ulcerans_Agy99 GVELINQWYNNPDYLAIMRDCKYTAIGVWSENSLDRTVVVAVYGSPT---
Mvan_0567|M.vanbaalenii_PYR-1 GIELINRWYHDPVDFAVMSDCSNTRIGVWSENSVDRTVVVAVYGKPQ---
Mflv_0523|M.gilvum_PYR-GCK GIELINQWYHDPVDLAVMSDCSNTRIGVWSENSVDRTVVVAVYGKPQ---
.::::* ** * *:: ** * :****:** **:*:*****.*
Rv1975|M.tuberculosis_H37Rv AMPPRGAVTGPPSPVAAQENVPIDPSPDYDASDEIEYGINWLPWILRGVY
MAV_2072|M.avium_104 --PARR--------------------------------------------
TH_0483|M.thermoresistible__bu --PVDGAPPAAGGGETGLHH------------------------------
MSMEG_0355|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4048|M.marinum_M --------------------------------------------------
MUL_3914|M.ulcerans_Agy99 --------------------------------------------------
Mvan_0567|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0523|M.gilvum_PYR-GCK --------------------------------------------------
Rv1975|M.tuberculosis_H37Rv PPPAMPPQ
MAV_2072|M.avium_104 --------
TH_0483|M.thermoresistible__bu --------
MSMEG_0355|M.smegmatis_MC2_155 --------
MMAR_4048|M.marinum_M --------
MUL_3914|M.ulcerans_Agy99 --------
Mvan_0567|M.vanbaalenii_PYR-1 --------
Mflv_0523|M.gilvum_PYR-GCK --------