For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGSTALNAARRTAELSALGDGVTPDVVVIGGGITGAGIALDAATRGLSVVLVEKHDLAFGTSRWSSKLVH GGLRYLASGNVGIARRSAIERGILMTRNAPHLVHAMPQLVPLLPEMNAASRALVRFGFAAGDGLRRLAGT PAETLPRSRRVDVQRAVSLAPTVRRRGLDGALLAYDGQLIDDARLVTAVARTAAQHGARVLTRVAASRAS RNSVTLTDTLTGESMQVSARAVVNATGVWAGEIDDSIKLRPSRGTHLVFGAEAFGNPTAALTVPIPGSLN RFVFAMPEQLGRVYLGLTDEDAPGPVPDVPEPTPAEITFLLDTVNTALDVALGPDDVVGAYAGLRPLIDA GGATADLSREHAVTESPSGVISVIGGKLTEYRYMAEDVVDRAVRLRGLTAGACRTRNLPLVGAPSNPVAT LRSPVDLPGSLVARYGAEAPNVIARASCERPTDQVADGIDVIRAEFEYAVTHEGALTVDDILDRRTRIGL VARDRDSAESTAKQIAEIAFQQGISE*
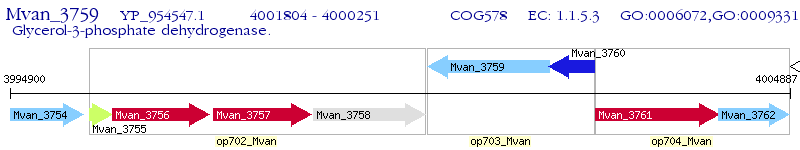
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3759 | - | - | 100% (517) | FAD dependent oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_1620 | - | 2e-65 | 36.64% (535) | FAD dependent oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2273c | glpD1 | 0.0 | 76.67% (510) | glycerol-3-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2774 | - | 0.0 | 87.48% (519) | FAD dependent oxidoreductase |
| M. tuberculosis H37Rv | Rv2249c | glpD1 | 0.0 | 76.67% (510) | glycerol-3-phosphate dehydrogenase |
| M. leprae Br4923 | MLBr_00713 | glpD | 6e-65 | 37.10% (531) | putative glycerol-3-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3882c | - | 1e-158 | 57.53% (511) | glycerol-3-phosphate dehydrogenase 1 GlpD1 |
| M. marinum M | MMAR_3342 | glpD1 | 0.0 | 76.68% (506) | glycerol-3-phosphate dehydrogenase GlpD1 |
| M. avium 104 | MAV_2188 | - | 0.0 | 77.89% (502) | glycerol-3-phosphate dehydrogenase 1 |
| M. smegmatis MC2 155 | MSMEG_4332 | - | 0.0 | 78.53% (503) | glycerol-3-phosphate dehydrogenase 1 |
| M. thermoresistible (build 8) | TH_4606 | - | 0.0 | 77.52% (516) | PUTATIVE FAD dependent oxidoreductase |
| M. ulcerans Agy99 | MUL_1300 | glpD1 | 0.0 | 76.28% (506) | glycerol-3-phosphate dehydrogenase GlpD1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3759|M.vanbaalenii_PYR-1 ------------MSGSTALNAARRTAELSALGDGVTPDVVVIGGGITGAG
Mflv_2774|M.gilvum_PYR-GCK ------------MSGSTSLNAARRTVDLTQLGDGAPVDVVVIGGGITGAG
TH_4606|M.thermoresistible__bu --------VSAKKGDATALNASRRAAELTALAEQR-LDILVIGGGITGAG
Mb2273c|M.bovis_AF2122/97 ----------MLMPHSAALNAARRSADLTALADGGALDVIVIGGGITGVG
Rv2249c|M.tuberculosis_H37Rv ----------MLMPHSAALNAARRSADLTALADGGALDVIVIGGGITGVG
MMAR_3342|M.marinum_M ------------MPDSTALNAARRSTELTALADGEALDVLVIGGGITGVG
MUL_1300|M.ulcerans_Agy99 ------------MPDSTALNAARRSTELTALADGEALDVLVIGGGITGVG
MAV_2188|M.avium_104 -----------MIPDPSALNAARRTADLAALAEGEPLDVLVIGGGITGAG
MSMEG_4332|M.smegmatis_MC2_155 ------MTASQGAASRTALNAERRAAELDTLAAGEHVDVLVIGGGITGAG
MAB_3882c|M.abscessus_ATCC_199 --------------MTTQLNSTRRSADLDALSGGAPIDVLVIGGGITGVG
MLBr_00713|M.leprae_Br4923 MTDLIQAPESEQTWPASALGPQQRAAAWERFG-TEQFDVVVIGGGVVGSG
: *.. :*:. :. *::*****:.* *
Mvan_3759|M.vanbaalenii_PYR-1 IALDAATRGLSVVLVEKHDLAFGTSRWSSKLVHGGLRYLASGNVGIARRS
Mflv_2774|M.gilvum_PYR-GCK IALDAASRGLAVVLVEKHDLAFGTSRWSSKLVHGGLRYLATGNVGIARRS
TH_4606|M.thermoresistible__bu IALDAASRGLTVALVEKHDLAFGTSRWSSKLVHGGLRYLASGNIGIARRS
Mb2273c|M.bovis_AF2122/97 IALDAATRGLTVALVEKHDLAFGTSRWSSKLVHGGLRYLASGNVGIARRS
Rv2249c|M.tuberculosis_H37Rv IALDAATRGLTVALVEKHDLAFGTSRWSSKLVHGGLRYLASGNVGIARRS
MMAR_3342|M.marinum_M IALDAASRGLQVALVEKHDLAFGTSRWSSKLVHGGLRYLASGNVGIARRS
MUL_1300|M.ulcerans_Agy99 IALDAASRGLQVALVEKHDLAFGTSRWSSKLVHGGLRYLASGNVGIARRS
MAV_2188|M.avium_104 IALDAAARGLRVALVEKRDLAFGTSRWSSKLVHGGLRYLASGNVGIARRS
MSMEG_4332|M.smegmatis_MC2_155 IALDAATRGLRVALVEKHDLAFGTSRWSSKLVHGGLRYLATGNVGIARRS
MAB_3882c|M.abscessus_ATCC_199 VALDAASRGLRVTLVEKHDLAFGTSRFSSKLVHGGLRYLASGQVGVARES
MLBr_00713|M.leprae_Br4923 CALDAATRGLKVALVEARDLASGTSSRSSKMFHGGLRYLEQLEFGLVREA
*****:*** *.*** :*** *** ***:.******* :.*:.*.:
Mvan_3759|M.vanbaalenii_PYR-1 AIERGILMTRNAPHLVHAMPQLVPLLPEMNAASRALVRFGFAAGDGLRRL
Mflv_2774|M.gilvum_PYR-GCK ALERGILMTRNAPHLVHAMPQLVPLLPSMGAPSRALVRFGFAAGDGLRKL
TH_4606|M.thermoresistible__bu AVERGILMTRNAPHLVKAMPQLVPLIPTIGTLERALVRTGFLAGDVLRRL
Mb2273c|M.bovis_AF2122/97 AVERGILMTRNAPHLVHAMPQLVPLLPSMGHTKRALVRAGFLAGDALRVL
Rv2249c|M.tuberculosis_H37Rv AVERGILMTRNAPHLVHAMPQLVPLLPSMGHTKRALVRAGFLAGDALRVL
MMAR_3342|M.marinum_M AIERGILMTRNAPHLVHAMPQLVPLLPSMDHLKRALVRTGFLAGDALRML
MUL_1300|M.ulcerans_Agy99 AIERGILMTRNAPHLVHAMPQLVPLLPSMDHLKRALVRTGFLAGDALRML
MAV_2188|M.avium_104 ALERGILMTRNAPHLVHAMPQLVPLLPSMSRATRALVRGGFLAGDALRFL
MSMEG_4332|M.smegmatis_MC2_155 AVERGILMGRNAPHLVKAMPQLVPLLPSMNRASRALIRTGFLAGDGLRKL
MAB_3882c|M.abscessus_ATCC_199 AVERHILMTRTAPHLIHPLAQVVPLHPAVSMFSRTLVRVGFIAGDGLRKL
MLBr_00713|M.leprae_Br4923 LYERELSLTTLAPHLVKPLPFLFPLT--KRWWERPYIAAGIFLYDRLG--
** : : ****::.:. :.** *. : *: * *
Mvan_3759|M.vanbaalenii_PYR-1 AGTPAETLPRSRRVDVQRAVSLAPTVRRRGLDGALLAYDGQLIDDARLVT
Mflv_2774|M.gilvum_PYR-GCK AGTPASVLPRSRRVDAQRAVALAPTVRRDGLDGGLLAYDGQLIDDARLVT
TH_4606|M.thermoresistible__bu AGTPASTLPRSGRVDAGRAAELVPAVRRDRLDGALLAYDGQLIDDARLVT
Mb2273c|M.bovis_AF2122/97 AGTPAATLPRSRRIPASRVVEIAPTVRRDGLDGGLLAYDGQLIDDARLVM
Rv2249c|M.tuberculosis_H37Rv AGTPAATLPRSRRIPASRVVEIAPTVRRDGLDGGLLAYDGQLIDDARLVM
MMAR_3342|M.marinum_M AGTPSSTLPRSRRVSAQRVVELAPTVRRDGLAGGLLAYDGQLIDDARLVA
MUL_1300|M.ulcerans_Agy99 AGTPSSTLPRSRRVSAQRVVELAPTVRRDGLAGGLLAYDGQLIDDARLVT
MAV_2188|M.avium_104 AGTPASTLPRSQRVSAQRVVQLAPTVCRDGLDGGLLAYDGQLIDDARLVT
MSMEG_4332|M.smegmatis_MC2_155 AGTSAATLPRSRRIDAARAVAMAPTVRTDGLEGGFLAYDGQLVDDARLVV
MAB_3882c|M.abscessus_ATCC_199 AHTSAEVLPRSRAINAGEVIRRAPTVRRNGLRGGLMAFDGQLVDDARLVV
MLBr_00713|M.leprae_Br4923 ---GAKSVPAQKHLTRAGALRLSPGLKRSSLIGGIRYYD-TVVDDARHTL
: :* . : . * : * *.: :* ::**** .
Mvan_3759|M.vanbaalenii_PYR-1 AVARTAAQHGARVLTRVAASRASRNS-----VTLTDTLTGESMQVSARAV
Mflv_2774|M.gilvum_PYR-GCK AVARTAAQHGARILTRVAASDASRTA-----VSLTDTLTGETITIAARAV
TH_4606|M.thermoresistible__bu AVARTAAQHGARILTRVAADRARGDG-----AALTDQLTGESFDVAARAV
Mb2273c|M.bovis_AF2122/97 AVARTAAQHGARILTYVGASNVTGTS-----VELTDRRTRQSFALSARAV
Rv2249c|M.tuberculosis_H37Rv AVARTAAQHGARILTYVGASNVTGTS-----VELTDRRTRQSFALSARAV
MMAR_3342|M.marinum_M AVARTAAQYGARILTYVGASEATGTS-----ARLTDQRSGESFDVSARAV
MUL_1300|M.ulcerans_Agy99 AVARTAAQHGARILTYVGASEATGTS-----ARLTDQRSGESFDVSARAV
MAV_2188|M.avium_104 AVARTAAQHGARILTYVAASRATGTS-----VRLTDVRGGGSFDVSAGAV
MSMEG_4332|M.smegmatis_MC2_155 AVARTAAQHGARILTRVSADHATEES-----VRLTDHRTGSSFDVRARVV
MAB_3882c|M.abscessus_ATCC_199 SIARTAAGLGARILTRVAAEQVTGDG-----AVLRDRLTGNAIRVRAGLV
MLBr_00713|M.leprae_Br4923 TVARTAAHYGAVVRCSTQVVALLREGDRVIGVRVRDSEDGAVTEIRGHVV
::***** ** : . . . . : * : . *
Mvan_3759|M.vanbaalenii_PYR-1 VNATGVWAGEIDDSIKLRPSRGTHLVFGAEAFGNPTAALTVPIPGSLNRF
Mflv_2774|M.gilvum_PYR-GCK VNATGVWAGEIDESLTLRPSRGTHLVFDAAALGNPTAALTVPIPGEINRF
TH_4606|M.thermoresistible__bu INATGVWAGQVDPALRLRPSRGTHLVFDAAAFGNPVAALTIPIPGELNRF
Mb2273c|M.bovis_AF2122/97 INAAGVWAGEIDPSLRLRPSRGTHLVFDAKSFANPTAALTIPIPGELNRF
Rv2249c|M.tuberculosis_H37Rv INAAGVWAGEIDPSLRLRPSRGTHLVFDAKSFANPTAALTIPIPGELNRF
MMAR_3342|M.marinum_M INAAGVWGGQIDPALKLRPSRGTHLVFDANAFGNPTAALTIPIPGELNRF
MUL_1300|M.ulcerans_Agy99 INAAGVWGGQIDPALKLRPSRGTHLVFDANAFGNPTAALTIPIPGELNRF
MAV_2188|M.avium_104 INAAGVWAGEIDGSLRLRPSRGTHLVFDAAAFGNPTAALTIPIPGELNRF
MSMEG_4332|M.smegmatis_MC2_155 INAAGVWAGQVDPTIRLRPSRGTHLVFDAAAFGNPTAALTIPIPGELNRF
MAB_3882c|M.abscessus_ATCC_199 INAAGVWAGQVDREIKLRPSRGTHLVFRAASFGGLSAALTVPVPGTINRF
MLBr_00713|M.leprae_Br4923 VNATGVWTDEIQALSKQRGRFQVRVSKGVHVVVPRDRIVSDVAMILRTKK
:**:*** .::: * .:: . . :: .:
Mvan_3759|M.vanbaalenii_PYR-1 VFAMPEQLGRVYLGLTDEDAPGPVPDVPEPTPAEITFLLDTVNTALDVAL
Mflv_2774|M.gilvum_PYR-GCK VFAMPEQLGRVYLGLTDEDAPGPIPDVPEPTPAEVTFLLDTVNTALDTAL
TH_4606|M.thermoresistible__bu VFAMPEQLGRVYLGLTDEDAPGPIPDVPQPTPQEVSFLLDTVNTALATAL
Mb2273c|M.bovis_AF2122/97 VFAMPEQLGRIYLGLTDEDAPGPIPDVPQPSSEEITFLLDTVNTALGTAV
Rv2249c|M.tuberculosis_H37Rv VFAMPEQLGRIYLGLTDEDAPGPIPDVPQPSSEEITFLLDTVNTALGTAV
MMAR_3342|M.marinum_M VFAMPEQLGRVYLGLTDEDAPGPIPDVPEPSSEEIMFLLDTVNTALGTAL
MUL_1300|M.ulcerans_Agy99 VFAMPEQLGRVYLGLTDEDAPGPFLDVPEPSSEEIMFLLDTVNTALGTAL
MAV_2188|M.avium_104 VFAMPEQLGRVYLGLTDEAAPGPIPDVPEPSVEEITFLLDTVNTALGTAL
MSMEG_4332|M.smegmatis_MC2_155 VFAMPEQLGRIYLGLTDEDAPGPIPDVPQATPAEISFLLDTVNTALATTL
MAB_3882c|M.abscessus_ATCC_199 VFALPAIHDRVYLGLTDEPAPGPVPDVAEPSAAEERFLLDTANTVLQTSL
MLBr_00713|M.leprae_Br4923 SVMFIIPWGNHWIIGTTDTDWNLDLAHPAATKADIDYILQTVNTVLATPL
. : .. :: * : . . .: : ::*:*.**.* ..:
Mvan_3759|M.vanbaalenii_PYR-1 GPDDVVGAYAGLRPLIDAG-----GATADLSREHAVTESPSGVISVIGGK
Mflv_2774|M.gilvum_PYR-GCK RHEDVVGSYAGLRPLIDTGT----GRTADVSREHAVTESPSGVISVIGGK
TH_4606|M.thermoresistible__bu GPDDVIGAYAGLRPLIDTPG----GRTADISRDHAVVESDTGLFSVLGGK
Mb2273c|M.bovis_AF2122/97 GTKDVIGAYAGLRPLIDTGGAGVQGRTADVSRDHAVFESPSGVISVVGGK
Rv2249c|M.tuberculosis_H37Rv GTKDVIGAYAGLRPLIDTGGAGVQGRTADVSRDHAVFESPSGVISVVGGK
MMAR_3342|M.marinum_M QTSDVIGAYAGLRPLIDTG----EGRTADVSRDHAVIESPSGVISVVGGK
MUL_1300|M.ulcerans_Agy99 QTSDVIGAYAGLRPLIDTG----EGRTADVSRDHAVIESPSGVISVVGGK
MAV_2188|M.avium_104 AATDVIGAYAGLRPLIDTG----EGRTADVSREHAVVESASGVISVIGGK
MSMEG_4332|M.smegmatis_MC2_155 TPADVRGTYAGLRPLIDAGE----GRTADLSREHAVVESPDGVLSVVGGK
MAB_3882c|M.abscessus_ATCC_199 SETDVVGRFAGLRPLLDGDD----SATADLSRKHAVRVSESGVVSVVGGK
MLBr_00713|M.leprae_Br4923 THADIDGVYAGLRPLLAGES----DDTSKLTREHAVAVPVAGLVAIAGGK
*: * :******: . *:.::*.*** . *:.:: ***
Mvan_3759|M.vanbaalenii_PYR-1 LTEYRYMAEDVVDRAVRLRGLTAGACRTRNLPLVGAPSNPVATLRSPVD-
Mflv_2774|M.gilvum_PYR-GCK LTEYRYMAEDVVDRAVELRGLTAGVCRTRNLPLVGAPSNPVATLRSPVE-
TH_4606|M.thermoresistible__bu LTEYRHMAEDAVDRAVTSRGLRAGECRTRNLPLVGAPANPVATFRSAVDS
Mb2273c|M.bovis_AF2122/97 LTEYRYMAEDVLNRAITLRHLRAAKCRTRNLPLIGAPANPGPAPGSGAG-
Rv2249c|M.tuberculosis_H37Rv LTEYRYMAEDVLNRAITLRHLRAAKCRTRNLPLIGAPANPGPAPGSGAG-
MMAR_3342|M.marinum_M LTEYRYMAEDVLNRAISLRQLQANQCRTRDLPLIGAPANPGPAAGPG-G-
MUL_1300|M.ulcerans_Agy99 LTEYRYMAEDVLNRAISLRQLQATQCRTRDLPLIGAPATPGPAAGPG-G-
MAV_2188|M.avium_104 LTEYRYMAQDVLDRAVRVRRLRAAKCRTRNLPLIGAPANPGITGAPQVA-
MSMEG_4332|M.smegmatis_MC2_155 LTEYRYMAEDVLDRAVALRGLDAGECRTRNLPLVGAPANPVTTLRAQVE-
MAB_3882c|M.abscessus_ATCC_199 LTTYRKMAQDALDAGLQRRPLAASECVTQNLAVVDDWP------------
MLBr_00713|M.leprae_Br4923 YTTYRVMAADAIDAAVAFVPARVAPSITEKVGLLGADGYFALINQVEHVA
* ** ** *.:: .: . . *..: ::.
Mvan_3759|M.vanbaalenii_PYR-1 --------LPGSLVARYGAEAPNVIARASCE-RPTDQVADGIDVIRAEFE
Mflv_2774|M.gilvum_PYR-GCK --------LPGSLVARFGAEAPNVIARASCA-RPTDPVAEGIDVVRAEFE
TH_4606|M.thermoresistible__bu GAE-LPARLPSSLVARYGAEAPNVIARAACD-RPTDPVAEGIDVIRAEFE
Mb2273c|M.bovis_AF2122/97 --------LPESLVARYGAEAANVAAAATCE-RPTEPVADGIDVTRAEFE
Rv2249c|M.tuberculosis_H37Rv --------LPESLVARYGAEAANVAAAATCE-RPTEPVADGIDVTRAEFE
MMAR_3342|M.marinum_M --------LPESLVARFGAEAANVVATATCE-RPTDPVAEGIDVTRAEFE
MUL_1300|M.ulcerans_Agy99 --------LPEFLVARFGAEAANVVVTATCE-RPTDPVAEGIDVTRAEFE
MAV_2188|M.avium_104 --------LPESLVSRYGAEAAKVVATAGCE-RPSEPVVDGIDVTRAEFE
MSMEG_4332|M.smegmatis_MC2_155 --------LPGSLVARYGAEAHNVIAQATCA-RPVEPVADGIDVTRAEFE
MAB_3882c|M.abscessus_ATCC_199 -----------------GAAGAG--------------LVEGVDITREQVE
MLBr_00713|M.leprae_Br4923 ALQGLHPYRVRHLLDRYGALIGDVLALAAEAPDLLSPIQEAPGYLKVEAR
** : :. . : : .
Mvan_3759|M.vanbaalenii_PYR-1 YAVTHEGALTVDDILDRRTRIGLVARDR-----DSAESTAKQI-AEIAFQ
Mflv_2774|M.gilvum_PYR-GCK YAVTHEGALTVDDILDRRTRIGLVARDR-----VRAESVADEILAEIAFQ
TH_4606|M.thermoresistible__bu YAVTHEGAMTVEDILDRRTRIGLVAADR-----QRAVAAAEEILAAAE--
Mb2273c|M.bovis_AF2122/97 YAVTHEGALDVDDILDRRTRIGLVPRDR-----ERVVAVAKEFLSR----
Rv2249c|M.tuberculosis_H37Rv YAVTHEGALDVDDILDRRTRIGLVPRDR-----ERVVAVAKEFLSR----
MMAR_3342|M.marinum_M YAVTHEGALDAEDILDRRTRIGLVPHDR-----QRVVGIAEEFLAGVG--
MUL_1300|M.ulcerans_Agy99 YAVTHEGALDAEDILDRRTRIGLVPHDR-----ERVVGIAEEFLAGVG--
MAV_2188|M.avium_104 YAITHEGALEVSDIVDRRTRIGLVPRDR-----ERVVAVAEEFLARFG--
MSMEG_4332|M.smegmatis_MC2_155 YAVTHEGAMTVDDILDRRTRIGLVTEDR-----ERALAAATEVLARIG--
MAB_3882c|M.abscessus_ATCC_199 FALTHEGALDADDILDRRTRIGLVPVDK-----NQAIGAVQRLVDSYFSQ
MLBr_00713|M.leprae_Br4923 YAVTAEGALHLEDILARRMRVSIEYPHRGVACAREVADVVAPVLGWTAED
:*:* ***: .**: ** *:.: .: . . .
Mvan_3759|M.vanbaalenii_PYR-1 QGISE-------------------------------------------
Mflv_2774|M.gilvum_PYR-GCK QGISE-------------------------------------------
TH_4606|M.thermoresistible__bu ------------------------------------------------
Mb2273c|M.bovis_AF2122/97 ------------------------------------------------
Rv2249c|M.tuberculosis_H37Rv ------------------------------------------------
MMAR_3342|M.marinum_M ------------------------------------------------
MUL_1300|M.ulcerans_Agy99 ------------------------------------------------
MAV_2188|M.avium_104 ------------------------------------------------
MSMEG_4332|M.smegmatis_MC2_155 ------------------------------------------------
MAB_3882c|M.abscessus_ATCC_199 VN----------------------------------------------
MLBr_00713|M.leprae_Br4923 IDREVATYNARVEAEVLSQAQPDDVSADMLRASAPEARTKIIEPVSLT