For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LVHHRIARPDAAADRGATDVTEIIAFGEFGALMDMASCRSDPFAIDFEVLSAPELDDLNDLADESPRLLL APTGSTNTAPPTVPIHVGGAHLRECPRPSRRALSGSNVRASGGAHRKTPEAAGAAHSRLVITAIAVGAIA AAANAADDATEAQQPVLVADTAASPAALPGPSGMQIVPVANSFDAGIHRQELANGTAYAQERAQREARLR RPQFAFPSRGILTSGFGARWGTLHAGLDIANAVGTPIYAASDGEVIASGPTPGFGMWVKIRAADGTVTLY GHIDTTNVQTGQRVMAGDQIATMGNRGNSTGPHLHFEVHLNGTDKIDPMAWLGERGIPND
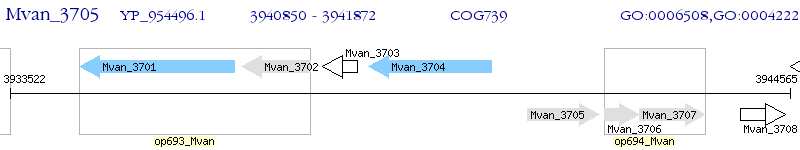
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3705 | - | - | 100% (340) | peptidase M23B |
| M. vanbaalenii PYR-1 | Mvan_4871 | - | 6e-80 | 48.55% (346) | peptidase M23B |
| M. vanbaalenii PYR-1 | Mvan_2208 | - | 1e-05 | 30.97% (113) | peptidase M23B |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0975c | - | 6e-77 | 46.61% (339) | hypothetical protein Mb0975c |
| M. gilvum PYR-GCK | Mflv_2817 | - | 1e-168 | 86.14% (339) | peptidase M23B |
| M. tuberculosis H37Rv | Rv0950c | - | 6e-77 | 46.61% (339) | hypothetical protein Rv0950c |
| M. leprae Br4923 | MLBr_00154 | - | 9e-72 | 46.57% (335) | hypothetical protein MLBr_00154 |
| M. abscessus ATCC 19977 | MAB_1055c | - | 2e-65 | 54.74% (232) | hypothetical protein MAB_1055c |
| M. marinum M | MMAR_4551 | - | 1e-75 | 46.59% (367) | hypothetical protein MMAR_4551 |
| M. avium 104 | MAV_1073 | - | 1e-77 | 47.88% (353) | M23 peptidase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_5526 | - | 9e-77 | 47.26% (347) | peptidoglycan-binding LysM |
| M. thermoresistible (build 8) | TH_1774 | - | 4e-77 | 47.71% (350) | peptidoglycan-binding LysM |
| M. ulcerans Agy99 | MUL_4721 | - | 4e-75 | 46.41% (362) | hypothetical protein MUL_4721 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0975c|M.bovis_AF2122/97 ------------MAAIRTPRDRWPHHHRNEVTEIIPLDGFLDGLALYDEL
Rv0950c|M.tuberculosis_H37Rv ------------MAAIRTPRDRWPHHHRNEVTEIIPLDGFLDGLALYDEL
MMAR_4551|M.marinum_M -MSQHRYARTAVVGDPRTSRDRWLQHHRNEVTEIIPLDGFDE----LDEL
MUL_4721|M.ulcerans_Agy99 -MSQHRYARTAVVGNPRTSRDRWLQHHRNEVTEIIPLDGF-------DEL
MLBr_00154|M.leprae_Br4923 ----------MAAEAVRSFRNRWLNHQRNDVTEIIPVDGFDD-FDDLYDL
MAV_1073|M.avium_104 ------------MRVGRAPGARSAEPHRTEVTEILPLDGFD-----FDDL
MSMEG_5526|M.smegmatis_MC2_155 ------------------------------MTDIIPFSEFSKAFDQFAAL
TH_1774|M.thermoresistible__bu LAKHRAVSPAGDSTDAHRHPRQGFDPHPAEITEIIPLSGFGRLSDLGADT
MAB_1055c|M.abscessus_ATCC_199 ----------------------------------MPIDEFG----LSDAL
Mvan_3705|M.vanbaalenii_PYR-1 -----------MVHHRIARPDAAADRGATDVTEIIAFGEFG------ALM
Mflv_2817|M.gilvum_PYR-GCK -----------MVHHRLARPDAPADRGATDVTEIIAFGEFG------ALL
:... *
Mb0975c|M.bovis_AF2122/97 DFAELD----DLDLGDDC-VFDYEAQLLAAPELDDLD---DADDLAPEWL
Rv0950c|M.tuberculosis_H37Rv DFAELD----DLDLGDDC-VFDYEAQLLAAPELDDLD---DADDLAPEWL
MMAR_4551|M.marinum_M ELADLD----DLDFGDP--EFDTGRQLLQAPELDDLD---EIDNLIPAWL
MUL_4721|M.ulcerans_Agy99 ELADLD----DLDFGDP--EFDTGRQLLQAPELDDLD---EIDNLIPAWL
MLBr_00154|M.leprae_Br4923 DFAEID----ELQFSDDDYNFCNETQVLLASEIDDLQ---DIDSLASEPL
MAV_1073|M.avium_104 DFCDDAGDHGDLDFSTDS-PFDNEAQVLLAPELDDLN---ETDDLRPLWL
MSMEG_5526|M.smegmatis_MC2_155 DTQISS------AYDNLS-AFEREARVIEAPELDDLN---DTDDLEPVRL
TH_1774|M.thermoresistible__bu DFYGDA------WDDEWDVAFDRESEVVSAPELDDL----DPDAEAPLRL
MAB_1055c|M.abscessus_ATCC_199 ERPELE------DYDVYEDAPEEGAEIEDQTALDQLEQDWEEDTDEADRI
Mvan_3705|M.vanbaalenii_PYR-1 DMASCR-----------SDPFAIDFEVLSAPELDDLN---DLADESPRLL
Mflv_2817|M.gilvum_PYR-GCK DMANCR-----------SDPFAMDFEVLSAPELDDLN---DVADESPRLL
: .: . :*:* : . :
Mb0975c|M.bovis_AF2122/97 VAP-TVVLT----------------------------------PEVTPVS
Rv0950c|M.tuberculosis_H37Rv VAP-TVVLT----------------------------------PEVTPVS
MMAR_4551|M.marinum_M ATP-SPVLVGATSE-----------------ALPEPSAAVPPAANVAPAP
MUL_4721|M.ulcerans_Agy99 ATP-SPVLVGATSE-----------------ALPEPSAAVPPAANVAPAP
MLBr_00154|M.leprae_Br4923 AAS-ANVAP-----------------------------------NFTSAP
MAV_1073|M.avium_104 AAPVTEVLPRTTVA-----------------ARTERRPERPRGADVVGVA
MSMEG_5526|M.smegmatis_MC2_155 DIP-SEFLADARDAGERSASRRKRNATHAYRDSHTDVSDGTAATDVIDMT
TH_1774|M.thermoresistible__bu VVP-SEFRPDPRDE---------RPSGHAYRDSHTDVSDGTAATDVIAI-
MAB_1055c|M.abscessus_ATCC_199 RAS-IEATAD-------------WFAASAART--RDPRD--TPTDKLPRI
Mvan_3705|M.vanbaalenii_PYR-1 LAPTGSTNTAPPTVP----------IHVGGAHLRECPRPSRRALSGSNVR
Mflv_2817|M.gilvum_PYR-GCK LPPKGATKAASPTVP----------APVGGARLRERPPPSR-AQSGATLR
. .
Mb0975c|M.bovis_AF2122/97 RRVGQHRK--QPIGAARGRLLISAMAAGAAAAAAHTAIQQ-SETPRTETV
Rv0950c|M.tuberculosis_H37Rv RRVGQHRK--QPIGAARGRLLISAMAAGAAAAAAHTAIQQ-SETPRTETV
MMAR_4551|M.marinum_M RRGGQHRK--QSTSAAKGRLLIGSMAAGAAAAAAHTVTHQ-SDVTSMDTV
MUL_4721|M.ulcerans_Agy99 RRGGQHRK--QSTSAVKGRLLIGSMAAGAAAAAAHTVTHQ-SDVTSMDTV
MLBr_00154|M.leprae_Br4923 RRPCQHRK--QPTSATRGRLLISAMAAGAMATAAHTVISH-DDIPKTETV
MAV_1073|M.avium_104 RRGGQHRK--QPTSAARGRLLISAMAAGAAAAAAHTATSH-ADTPKTDAV
MSMEG_5526|M.smegmatis_MC2_155 GRRGAHRK--ETTGALKSRLMIAAVAAGATAAGAYSFGNG-TEAQAADDT
TH_1774|M.thermoresistible__bu GRRGQHRKP-EPTSAGKGRLMIAAMAAGATAAGAYSVVNGDDEKSSTDAT
MAB_1055c|M.abscessus_ATCC_199 SSGGVHRRR-EIGQVGKTRLALAAMAAGAAAAAGYNALSEHDAATAADHH
Mvan_3705|M.vanbaalenii_PYR-1 ASGGAHRKTPEAAGAAHSRLVITAIAVGAIAAAANAADDATEAQQPVLVA
Mflv_2817|M.gilvum_PYR-GCK ASGGAHRRTPAATGAVHSRLVITAIAVGAIAAAANAAVDETEAQQPVLVA
**: . : ** : ::*.** *:..
Mb0975c|M.bovis_AF2122/97 LTAHASALNEGSG-SNPPRGVQVIAAQPAASAAVHNAEFARGVAFAEERA
Rv0950c|M.tuberculosis_H37Rv LTAHASALNEGSG-SNPPRGVQVIAAQPAASAAVHNAEFARGVAFAEERA
MMAR_4551|M.marinum_M LTANASALIGGSD-SNEARGVQVIAAQPATNVEAHKAEFARGVAFAQERA
MUL_4721|M.ulcerans_Agy99 LTANASALIGGSD-SNEARGVQVIAAQPATNVEAHKAEFARGVAFAQERA
MLBr_00154|M.leprae_Br4923 LTANASALSGATV-SDATQGVQVITVHPAASTAVHNEEHAKGTAFAHERA
MAV_1073|M.avium_104 LTANASSLTGGAGGSNTVRGPQVIAAEPAATAALHNQELAKGVAFANDRA
MSMEG_5526|M.smegmatis_MC2_155 VLATGQTTVDGASVSGSNDGMQIVSVASNISSAVHAEEITKAAAFAQERA
TH_1774|M.thermoresistible__bu VLAADQAAMP-DAIDGATGGMQIVTVAQPAKAIVHAEEITKAAAFAQERA
MAB_1055c|M.abscessus_ATCC_199 GLALGN-----SAAVMATHGPQLVSAPLATDASVTDEQLAKATAFATERA
Mvan_3705|M.vanbaalenii_PYR-1 DTAASPAALPGPS------GMQIVPVANSFDAGIHRQELANGTAYAQERA
Mflv_2817|M.gilvum_PYR-GCK DSAASPTALPDAS------GMQIIPVANGFDAGIHRQELANGAAFAQERA
* * *::.. : :...*:* :**
Mb0975c|M.bovis_AF2122/97 EREARLQRPLYVMPTKGIFTSSFGYRWGVLHAGIDLANAIGTPIYAVSDG
Rv0950c|M.tuberculosis_H37Rv EREARLQRPLYVMPTKGIFTSSFGYRWGVLHAGIDLANAIGTPIYAVSDG
MMAR_4551|M.marinum_M QREARLQQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANSIGTPIYAVSDG
MUL_4721|M.ulcerans_Agy99 QREARLQQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANSIGTPIYAVSDG
MLBr_00154|M.leprae_Br4923 QREARLSQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANAIGTPILAVSDG
MAV_1073|M.avium_104 QREARLQQPLYVMPTKGIFTSNFGYRWGVLHAGIDLANSIGTPIYAVSDG
MSMEG_5526|M.smegmatis_MC2_155 EREARLSRPLFVMPTKGVWTSGFGYRWGVLHGGIDIANSIGTPIYAASDG
TH_1774|M.thermoresistible__bu EREARLARPQFVMPTTGVWTSGFGYRWGVLHGGIDIANSVGTPILAAADG
MAB_1055c|M.abscessus_ATCC_199 DREKRLLAPRFVMPTNGTFTSGFGYRWGALHGGIDIANSIGTPIVAAADG
Mvan_3705|M.vanbaalenii_PYR-1 QREARLRRPQFAFPSRGILTSGFGARWGTLHAGLDIANAVGTPIYAASDG
Mflv_2817|M.gilvum_PYR-GCK QREARLRRPQFTFPARGILTSGFGARWGTLHAGLDIANAVGTPIYAASDG
:** ** * :.:*: * **.** ***.**.*:*:**::**** *.:**
Mb0975c|M.bovis_AF2122/97 VVIDAGPTAGYGMWVKLLHADGTVTLYGHVNTTLVSVGERVMAGDQIATM
Rv0950c|M.tuberculosis_H37Rv VVIDAGPTAGYGMWVKLLHADGTVTLYGHVNTTLVSVGERVMAGDQIATM
MMAR_4551|M.marinum_M LVIDAGPTAGYGMWVKIRHADGTVTLYGHVNTTLVSVGQRVMAGDQIATM
MUL_4721|M.ulcerans_Agy99 LVIDAGPTAGYGMWVKIRHADGTVTLYGHVNTTLVNVGQRVMAGDHIATM
MLBr_00154|M.leprae_Br4923 VVIDVGPTAGYGMWVKLRHADGTVTLYGHVNTTLVNVGQYVVAGDQIATM
MAV_1073|M.avium_104 VVIEAGPAGGYGMLVKLRHADGTVTLYGHINTALVSVGERVMAGDQIATM
MSMEG_5526|M.smegmatis_MC2_155 VVTDVGPTAGYGAWVKIRHSDGTVTLYGHINTWLVSVGERVMAGDQIATM
TH_1774|M.thermoresistible__bu VVIETGPQGGYGIWVKLRHYDGTVTVYAHLSSVLVNVGQRVMAGDQIARM
MAB_1055c|M.abscessus_ATCC_199 VVIATGPTAGYGAWVKIRHSDGTVTLYGHINTWEVSVGQRVMAGDRIATI
Mvan_3705|M.vanbaalenii_PYR-1 EVIASGPTPGFGMWVKIRAADGTVTLYGHIDTTNVQTGQRVMAGDQIATM
Mflv_2817|M.gilvum_PYR-GCK DVIASGPTPGFGMWVKILAADGTVTLYGHIDTTMVQTGQRVMAGDQIATM
* ** *:* **: *****:*.*:.: *..*: *:***:** :
Mb0975c|M.bovis_AF2122/97 GSRGFSTGPHLHFEVLLGGTERVDPVPWLAKRGLSVGN-YTG
Rv0950c|M.tuberculosis_H37Rv GSRGFSTGPHLHFEVLLGGTERVDPVPWLAKRGLSVGN-YTG
MMAR_4551|M.marinum_M GNRGFSTGPHLHFEVLQGGTERIDPVPWLAKRGLNVGN-YAG
MUL_4721|M.ulcerans_Agy99 GNRGFSTGPHLHFEVLQGGTERIDPVPWLAKRGLNVGN-YAG
MLBr_00154|M.leprae_Br4923 GTRGNSTGPHLHFEVLLSGSERIDPVPWLAKRGIYVGN-YTG
MAV_1073|M.avium_104 GNRGNSTGPHLHFEVLQGGTERIDPVPWLAKRGLMVGN-YAG
MSMEG_5526|M.smegmatis_MC2_155 GNRGYSTGPHLHFEVLTNGTNRIDPVGWLAKRGLSPGS-YVG
TH_1774|M.thermoresistible__bu GNTGNSTGPHLHFEVLLDGVNRVDPAAWLAKRGVGPNSGYMR
MAB_1055c|M.abscessus_ATCC_199 GNRGNSTGPHLHFEVLLGGSQRIDPQGWLANKGLTFTR-FGD
Mvan_3705|M.vanbaalenii_PYR-1 GNRGNSTGPHLHFEVHLNGTDKIDPMAWLGERGIPND-----
Mflv_2817|M.gilvum_PYR-GCK GNRGNSTGPHLHFEVHRNGSVKTDPMAWLGERGVPNG-----
*. * ********** .* : ** **.::*: