For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTSAPPRTAPRTDSGCPAPASTGTGTSGFYRNDLDGLRGIAIALVAVFHVWFGRVSGGVDVFLALSGFFF GGRLLRAALTSTASLAPVPEVRRLIRRLFPALVVVLTACAGLTILVQPETRWETFADQSLASLGYYQNWE LSSTASNYLRAGEAVSPLQHIWSMSVQGQFYIAFLLLVFGMAFLLRGLLLGRHLRVLLIVLLSALTTASF VYAIVAHQADQTTAYYNSFARAWELLLGALAGALVHHVRWPMWLRTVVSVVALAAILSCGAFIDGVAEFP GPWALVPVGATILLILSAANRAADPHLARTNGRMPLPNRLLATKPFVSLGAMAYSLYLWHWPLLIFWLAY SGHTQANLAEGAGVLLVSGVLAWLTTRYIEDPLRLRATSSAKKAKATVPLGTRLRRPTIVLGLVVTLLGG ALTAASVTWREHIAVQRANGEELAGLSPRDYPGARALIDHAKVPKLPMRPTVLEAKDDLPQSTTDNCISD FADPSLRECAFGSKTATRTIAVAGGSHAEHWITALDLLGRLYGFKVVTYLKMGCPLSAERAPRIPGSGEL YPQCFDWVQAAMREIVSDRPDFVFTTVTRPRDAQPGDVMPDVYLPIWQAFAESGVKVLGMRDTPWLVRNG EPFFPSDCLADGGDATTCGMPRALVLDDVNPAADLTKRFPMMGMIDMSNALCRDDYCRAAEGNILIYRDS HHLSATYVRSMAPELGRRIGALTGWWP*
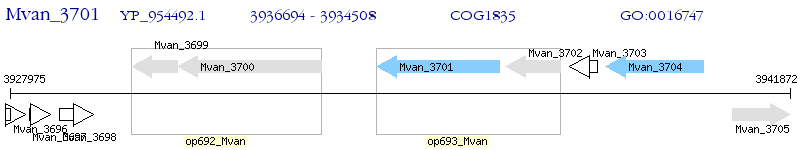
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3701 | - | - | 100% (728) | acyltransferase 3 |
| M. vanbaalenii PYR-1 | Mvan_2787 | - | 0.0 | 69.58% (733) | acyltransferase 3 |
| M. vanbaalenii PYR-1 | Mvan_1670 | - | 4e-40 | 30.59% (376) | acyltransferase 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1592c | - | 0.0 | 68.44% (732) | hypothetical protein Mb1592c |
| M. gilvum PYR-GCK | Mflv_2821 | - | 0.0 | 93.68% (728) | acyltransferase 3 |
| M. tuberculosis H37Rv | Rv1565c | - | 0.0 | 68.44% (732) | hypothetical protein Rv1565c |
| M. leprae Br4923 | MLBr_01213 | - | 0.0 | 64.66% (730) | hypothetical protein MLBr_01213 |
| M. abscessus ATCC 19977 | MAB_2689 | - | 0.0 | 62.01% (716) | acyltransferase |
| M. marinum M | MMAR_2380 | - | 0.0 | 67.82% (721) | hypothetical protein MMAR_2380 |
| M. avium 104 | MAV_3209 | - | 0.0 | 64.42% (728) | acyltransferase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3187 | - | 0.0 | 71.07% (726) | acyltransferase domain-containing protein |
| M. thermoresistible (build 8) | TH_1799 | - | 0.0 | 67.71% (706) | CONSERVED HYPOTHETICAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1556 | - | 0.0 | 67.55% (721) | hypothetical protein MUL_1556 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3701|M.vanbaalenii_PYR-1 ---------MLTSAPPRTAPRTDSGCPAPASTGTGTSG---FYRNDLDGL
Mflv_2821|M.gilvum_PYR-GCK ---------MLTSAPPRTAPRTDSGRPAPALTGTGAPG---FYRHDLDGL
MSMEG_3187|M.smegmatis_MC2_155 ----------MTLSPTRPAPISRDTRAANPSMGTRKSG---FYRHDLDGL
TH_1799|M.thermoresistible__bu ------------------------------VTGPGTSG---FYRYDLDGL
Mb1592c|M.bovis_AF2122/97 ---------MLTLSPPRPPALTPEPALPPVTMGTRTTG---FYRHDLDGL
Rv1565c|M.tuberculosis_H37Rv ---------MLTLSPPRPPALTPEPALPPVTMGTRTTG---FYRHDLDGL
MMAR_2380|M.marinum_M ---------MQTLTPPTP----PTAAAAPVAMGTRKTG---FYRHDLDGL
MUL_1556|M.ulcerans_Agy99 ---------MQTLTPPTS----PTAAAAPVAMGTRKTG---FYRHDLDGL
MLBr_01213|M.leprae_Br4923 MSSVNWRSIMQTLSPASL----LVATAKPVPLEGRTAVFTIFYRHDLDGL
MAV_3209|M.avium_104 ---------MQTPSPSPA----PVAAADPGTSGARTPGSRGFYRYDLDGL
MAB_2689|M.abscessus_ATCC_1997 ---------MFFVSATKPTKDPEVSPAAAMDATPKPKGDKAFYRYDLDGL
*** *****
Mvan_3701|M.vanbaalenii_PYR-1 RGIAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGRLLRAALTSTASLAP
Mflv_2821|M.gilvum_PYR-GCK RGIAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGRLLRAALTSTASLAP
MSMEG_3187|M.smegmatis_MC2_155 RGIAIMMVAVFHIWFGRVSGGVDVFLALSGFFFGGKILRTALDQSTPLRP
TH_1799|M.thermoresistible__bu RGVAILLVALFHVWFGRVSGGVDVFLALSGFFFGGRLLRAVLAPGASLKP
Mb1592c|M.bovis_AF2122/97 RGVAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGKILRAALNPDLSLSP
Rv1565c|M.tuberculosis_H37Rv RGVAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGKILRAALNPDLSLSP
MMAR_2380|M.marinum_M RGIAIALVAIFHVWFGRVSGGVDVFLALSGFFFGGKILRTALNPSLKLSP
MUL_1556|M.ulcerans_Agy99 RGIAIALVAIFHVWFGRVSGGVDVFLALSGFFFGGKILRTALNPSLKLSP
MLBr_01213|M.leprae_Br4923 RGIAIALVAIFHVWFGRVSGGVDVLLAMSGFFFGGKILRAALNPVPSLSP
MAV_3209|M.avium_104 RGIAIALVAVFHIWFGRVSGGVDVFLALSGFFFGGKVIRAALNPAVALSP
MAB_2689|M.abscessus_ATCC_1997 RGIAIFLVAVFHVWFGRVSGGVDVFLTLSGFFYGSKLLRTATTQGASLNP
**:** :**:**:***********:*::****:*.:::*:. * *
Mvan_3701|M.vanbaalenii_PYR-1 VPEVRRLIRRLFPALVVVLTACAGLTILVQPETRWETFADQSLASLGYYQ
Mflv_2821|M.gilvum_PYR-GCK VPEVRRLVRRLLPALVVVLAASAGLTILVQPETRWETFADQSLASLGYYQ
MSMEG_3187|M.smegmatis_MC2_155 LSEVVRLVRRLLPALVVVLAAAAVLTILIQPETRWEAFADQSLASLGYYQ
TH_1799|M.thermoresistible__bu LREVTRLVRRLLPALVVVLAASAVLTILIQPTTRWETFADQSLASLGYYQ
Mb1592c|M.bovis_AF2122/97 IAEVIRLIRRLLPALVVVLAGCALLTIAIQPQTRWEAFANQSLASLGYYQ
Rv1565c|M.tuberculosis_H37Rv IAEVIRLIRRLLPALVVVLAGCALLTIAIQPQTRWEAFANQSLASLGYYQ
MMAR_2380|M.marinum_M ITELIRLIRRLLPAMVVVLAACALLTILVQPQTRWEAFADQSLASLGYYQ
MUL_1556|M.ulcerans_Agy99 ITELIRLIRRLLPAMVVVLAACALLTILVQPQTRWEAFADQSLASLGYYQ
MLBr_01213|M.leprae_Br4923 VAEIIRLVRRLLPALVVVLTGCALLTVVMQPQTRWETFADQSLASLGYYQ
MAV_3209|M.avium_104 VAEVIRLVRRLVPALVVVLAGCAVLTILVQPETRWETFADQSLASLGYYQ
MAB_2689|M.abscessus_ATCC_1997 VPVVKRLVRRLLPALILVLAACAVLTVLVQPETRWETFAEQSLASLGYYQ
: : **:***.**:::**:..* **: :** ****:**:**********
Mvan_3701|M.vanbaalenii_PYR-1 NWELSSTASNYLRAGEAVSPLQHIWSMSVQGQFYIAFLLLVFGMAFLLRG
Mflv_2821|M.gilvum_PYR-GCK NWELSSTAANYLRASEAVSPLQHIWSMSVQGQFYLAFLLLVFGIGFLLRR
MSMEG_3187|M.smegmatis_MC2_155 NWELANTAADYLRAGETVSPLQHIWSMSVQGQFYIAFLVLIFGFAYLFRR
TH_1799|M.thermoresistible__bu NWELARTASDYLRAGEAVSPLQHIWSMSVQGQFYIAFLVLVMGFAVLFRR
Mb1592c|M.bovis_AF2122/97 NWELASTVSNYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLVAGCAYLLRR
Rv1565c|M.tuberculosis_H37Rv NWELASTVSNYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLVAGCAYLLRR
MMAR_2380|M.marinum_M NWELANSAADYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLIAGCTFLFRR
MUL_1556|M.ulcerans_Agy99 NWELANSAADYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLIAGCTFLFRR
MLBr_01213|M.leprae_Br4923 NWELVGTESSYLKAGEAVSPLQHIWSMSVQGQFDIAFLLLVAGCAYLFRR
MAV_3209|M.avium_104 NWELANTASDYLRAGEAVSPLQHIWSMSVQGQFYVSFLVLIAGLAYLFGR
MAB_2689|M.abscessus_ATCC_1997 NWELANTAADYLAASESVSPLQHLWSMSVQGQFYVGFLALVYLLAVLLRK
**** : :.** *.*:******:********* :.** *: *:
Mvan_3701|M.vanbaalenii_PYR-1 LLLG---RHLRVLLIVLLSALTTASFVYAIVAHQADQTTAYYNSFARAWE
Mflv_2821|M.gilvum_PYR-GCK LLVG---RHLRVLLIVLLSALTIASFVYAIGAHQADQTTAYYNSFARAWE
MSMEG_3187|M.smegmatis_MC2_155 VFG----RHLRTVFIVLLAALTIASFVYAIIAHNTDQATAYYNSFARAWE
TH_1799|M.thermoresistible__bu VLR----RHLRAAFVVLLTALTVASFVYAIIATNTDQATAYYNSFARAWE
Mb1592c|M.bovis_AF2122/97 LFRGPRAPYLRTMFVVLLSTLTLASFIYAIVAHHAYQATAYYNTFARAWE
Rv1565c|M.tuberculosis_H37Rv LFRGPRAPYLRTMFVVLLSTLTLASFIYAIVAHHAYQATAYYNTFARAWE
MMAR_2380|M.marinum_M LLG----THLRTMFVVLLSALTIASFVYAIIAHQAYQPTAYYNSFARAWE
MUL_1556|M.ulcerans_Agy99 LLG----THLRTMFVVLLSALTIASFVYAIIAHQAYQPTAYYNSFTRAWE
MLBr_01213|M.leprae_Br4923 PLG----TQLRIMFVVLLGALMIASFIYATFAHQANQTTAYYNSFARAWE
MAV_3209|M.avium_104 PLG----ARLRTLVLVVLAALTVASFGYAIVAHQQNQAAAYYDSFARGWE
MAB_2689|M.abscessus_ATCC_1997 PLG----RHIRTVLIVVIAVLSAASFGYAIYAHLDFQSIAYYNTFARAWE
: :* .:*:: .* *** ** * *. ***::*:*.**
Mvan_3701|M.vanbaalenii_PYR-1 LLLGALAGALVHHVRWPMWLRTVVSVVALAAILSCGAFIDGVAEFPGPWA
Mflv_2821|M.gilvum_PYR-GCK LLLGALAGALVQHVRWPMWLRTLVSVLALAAILSCGAFIDGVAEFPGPWA
MSMEG_3187|M.smegmatis_MC2_155 LLLGALAGALVGFVRWPMWLRTVVSVVSLAAILSCGWFIDGVKEFPGPWA
TH_1799|M.thermoresistible__bu LLLGALAGSLVPHIRWPKWLRALAATVALAAILSCGVLIDGVNEFPGPWA
Mb1592c|M.bovis_AF2122/97 LLAGALVGAVVPHVRWPMWLRTAVATAALAAILSCGALIDGVKEFPGPWA
Rv1565c|M.tuberculosis_H37Rv LLAGALVGAVVPHVRWPMWLRTAVATAALAAILSCGALIDGVKEFPGPWA
MMAR_2380|M.marinum_M LLLGALVGALVPYIRWPMWLRTIAATVGLAIVLSCGALINGVKEFPGPWA
MUL_1556|M.ulcerans_Agy99 LLLGALVGALVPYIRWPMWLRTIAATVGLAIVLSCGALINGVKEFPGPWA
MLBr_01213|M.leprae_Br4923 LLLGALVGAAVPYIRWPAWLRTVVATVALAAILSCGALINGVKEFPGPWA
MAV_3209|M.avium_104 LLLGALVGALVPRIRWPMWLRTLLATVALGAVLSCGALIDGVQEFPGPWA
MAB_2689|M.abscessus_ATCC_1997 LLLGVLVGALVAGTRWPRWLRQLLSFVAVVAILSCGALINGVREFPGPLA
** *.*.*: * *** *** : .: :**** :*:** ***** *
Mvan_3701|M.vanbaalenii_PYR-1 LVPVGATILLILSAANRAADPHLARTNGRMPLPNRLLATKPFVSLGAMAY
Mflv_2821|M.gilvum_PYR-GCK LVPVGATILLILSAANRAADPHLARADGRMPLPNRLLATKPFVSLGAMAY
MSMEG_3187|M.smegmatis_MC2_155 LVPVGATILFIFSAANRMSDPRTA---GRLPAPNRLLATAPFVSLGSMAY
TH_1799|M.thermoresistible__bu LVPVGATILFILSAAN------WQ---GPLPGPNRLLATTPFVTLGSIAY
Mb1592c|M.bovis_AF2122/97 LVPVGATMLMILAGANRQGHPGTR---DRLPLPNRLLATAPLVALGAMAY
Rv1565c|M.tuberculosis_H37Rv LVPVGATMLMILAGANRQGHPGTR---DRLPLPNRLLATAPLVALGAMAY
MMAR_2380|M.marinum_M LVPVGAAMLMIMAGANRQAHPSTR---DRLPLPNRLLAAGPLVTLGAMAY
MUL_1556|M.ulcerans_Agy99 LVPVGAAMLMIMAGANRQAHPSTR---DRLPLPNRLLAAGPLVTLGAMAY
MLBr_01213|M.leprae_Br4923 LVPVGAAMLLILAGANRQSRPGTS---ASMPLPNRLLATAPLVALGTIAY
MAV_3209|M.avium_104 LVPVGAAMLMILVGANLAG--------DRLPLPNRLLATRPLVELGAIAY
MAB_2689|M.abscessus_ATCC_1997 LVPVVATLLLILSAANLPAD-------ARQPVANRFLATRPLVELGALAY
**** *::*:*: .** * .**:**: *:* **::**
Mvan_3701|M.vanbaalenii_PYR-1 SLYLWHWPLLIFWLAYSGHTQANLAEGAGVLLVSGVLAWLTTRYIEDPLR
Mflv_2821|M.gilvum_PYR-GCK SLYLWHWPLLIFWLAYSGHAQANLAEGAGVLLVSGVLAWLTTRYIEVPLR
MSMEG_3187|M.smegmatis_MC2_155 SLYLWHWPLLIFWLSYSGHTAANFVEGAVILLVSGVLAWLTTRYIEEPLR
TH_1799|M.thermoresistible__bu SLYLWHWPLLIFWLAYTGNSGVNFVEGAVILLVSGVLAWLTTKLVEDPLR
Mb1592c|M.bovis_AF2122/97 SWYLWHWPLLIFWLSYTGHRHANFVEGAAVLLVSGLLAYLTTRLVEDPLR
Rv1565c|M.tuberculosis_H37Rv SWYLWHWPLLIFWLSYTGHRHANFVEGAAVLLVSGLLAYLTTRLVEDPLR
MMAR_2380|M.marinum_M SLYLWHWPLLIFWLSYTGHRHANFIEGAAILLVSGVLAFLTTRHVEDPLR
MUL_1556|M.ulcerans_Agy99 SLYLWHWPLLIFWLSYTGHRHANFIEGAAILLVSGVLAFLTTRHVEDPLR
MLBr_01213|M.leprae_Br4923 TLYLWHWPLLIFWLSYTGHHHANFVEGAALLLVSGLLAYLTTRLIENPLR
MAV_3209|M.avium_104 SLYLWHWPLLIFWLSYSGHKHAGFVEGTVLLLVSGVLAYLTNRLVEDPLR
MAB_2689|M.abscessus_ATCC_1997 SLYLWHWPLLVYWLVYTGEARVTVAEGAAILGISLALAYLTNKYIETPLR
: ********::** *:*. . . **: :* :* **:**.: :* ***
Mvan_3701|M.vanbaalenii_PYR-1 LRATS--SAKKAKATVPLGTRLRRPTIVLGLVVTLLGGALTAASVTWREH
Mflv_2821|M.gilvum_PYR-GCK LRATS--SAKKAKAAVPLRTRLLRPTIVLGSVVTLLGVALTAASFTWREH
MSMEG_3187|M.smegmatis_MC2_155 TQPGR--APVPA---VPLRARLRRPTIVLGSIVALLGVALTATSFTWREH
TH_1799|M.thermoresistible__bu YRKPA--ADVVVPVKPPLRTRLRRPTMVLGSVVALLGVALTATSFTWREH
Mb1592c|M.bovis_AF2122/97 YRAPAGVRSPAAVPPIPWRLRLRRPTIVLGSVVALLGVALTATSFTWREH
Rv1565c|M.tuberculosis_H37Rv YRAPAGVRSPAAVPPIPWRLRLRRPTIVLGSVVALLGVALTATSFTWREH
MMAR_2380|M.marinum_M YRAPSGTAAAVKTQAIPWQLRLRRPTIAMGSVVVLLGVTLTATSFTWREH
MUL_1556|M.ulcerans_Agy99 YRAPSGTAAAVKTQAIPWQLRLRRPTIAMGSVVVLLGVTLTATSFTWREH
MLBr_01213|M.leprae_Br4923 YRTLANTEYPSPARAAAWQLRLHRSTIALGSMVVLLGVALTATSFTWRQH
MAV_3209|M.avium_104 HRGSARPAPRPAAPTVPWLARVPRPTMALGSAIVLLGVTLTATSFTWRQH
MAB_2689|M.abscessus_ATCC_1997 Y-------PRVSPTSASLWTRLRRPTLALGTSIVLMAVALTATSFTWLEH
. *: *.*:.:* :.*:. :***:*.** :*
Mvan_3701|M.vanbaalenii_PYR-1 IAVQRANGEELAGLSPRDYPGARALIDHAKVPKLPMRPTVLEAKDDLPQS
Mflv_2821|M.gilvum_PYR-GCK VAVQRANGEELAGLSPRDYPGARALIDHAKVPKLPMRPTVLEAKDDLPQS
MSMEG_3187|M.smegmatis_MC2_155 VTVQRASGKELSGLSARDYPGARALIDHARVPKLPMRPTVLEAKNDLPIS
TH_1799|M.thermoresistible__bu VSLQRASGKELSGLAARDYPGARALTEGARVAKLPMRPSVHEAREDLPIS
Mb1592c|M.bovis_AF2122/97 VIVQRAAGKELSGLSSRDYPGARALIDHVRVPKLRMRPTVLEVRHDLPTS
Rv1565c|M.tuberculosis_H37Rv VIVQRAAGKELSGLSSRDYPGARALIDHVRVPKLRMRPTVLEVRHDLPTS
MMAR_2380|M.marinum_M VIVERASGKELRGLSANDYPGARALTDHVRVPKLRMRPTVFEAKGDFPAS
MUL_1556|M.ulcerans_Agy99 VIVERASGKELRGLSANDYPGARALTDHVRVPKLRMRPTVFEAKGDFPAS
MLBr_01213|M.leprae_Br4923 VIVLRATGKELSALNAQDYPGARALTAHARVPTLPMRPTVLEIKDDLPAS
MAV_3209|M.avium_104 VTVLRSAGKELSVLNPQDYPGARALTEHVRVPTLPMRPTVLEVKQDLPAS
MAB_2689|M.abscessus_ATCC_1997 VTVQRSNGKELSGLRPRDYPGAGALLYGDRVPDLPMRPTTLEASDDLPIT
: : *: *:** * ..***** ** :*. * ***:. * *:* :
Mvan_3701|M.vanbaalenii_PYR-1 TTDNCISDFADPSLRECAFGSKTATRTIAVAGGSHAEHWITALDLLGRLY
Mflv_2821|M.gilvum_PYR-GCK TADSCISDFADPSLRECAFGSKTATRTIAVAGGSHAEHWITALDLLGRLY
MSMEG_3187|M.smegmatis_MC2_155 TTEGCISDFANVGVINCTYGDKNATRTIALAGGSHAEHWITALDLLGQKH
TH_1799|M.thermoresistible__bu TIDGCISDFDDVSVIKCTYGDESASRTIALAGGSHAEHWITALDLLGRKH
Mb1592c|M.bovis_AF2122/97 TKDGCISDFVNPAIINCTYGDVDAPRTIALAGGSHAEHWLTALDLLGRMH
Rv1565c|M.tuberculosis_H37Rv TKDGCISDFVNPAIINCTYGDVDAPRTIALAGGSHAEHWLTALDLLGRMH
MMAR_2380|M.marinum_M TRDGCISDFVNPALVNCTYGDVDASRTIALAGGSHAEHWLPALDLLGRLH
MUL_1556|M.ulcerans_Agy99 TRDGCISDFVNPALVNCTYGDVDASRTIALAGGSHAEHWLPALDLLGRLH
MLBr_01213|M.leprae_Br4923 TRDGCISDFVNPAVVNCTYGDASANRTIALAGGSHAEHWLPALDVLGQLH
MAV_3209|M.avium_104 TRDGCISDFVNPAVVNCTYGDVTADRTIALAGGSHAEHWLPALDMLGKLH
MAB_2689|M.abscessus_ATCC_1997 TEQDCISDFRNRAVITCTYGNPHATRTIALAGGSHAEHWITALDILGRQH
* :.***** : .: *::*. * ****:*********:.***:**: :
Mvan_3701|M.vanbaalenii_PYR-1 GFKVVTYLKMGCPLSAERAPRIPGSGELYPQCFDWVQAAMREIVSDRPDF
Mflv_2821|M.gilvum_PYR-GCK GFKVVTYLKMGCPLSGERAPRIPGSGEPYPQCFDWVQAAMREIVADRPDF
MSMEG_3187|M.smegmatis_MC2_155 GFKVVTYLKMGCPLTTEEVPLVSGDNRPYPKCHEWNQRVMAKLIADHPDY
TH_1799|M.thermoresistible__bu HFKVVTYLKMGCPLTTDEMPRVSGDGRPYPKCRDWNERVLPELIADRPDY
Mb1592c|M.bovis_AF2122/97 HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCHQWVQAAMAKLVADHPDY
Rv1565c|M.tuberculosis_H37Rv HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCHQWVQAAMAKLVADHPDY
MMAR_2380|M.marinum_M HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCREWVQKTMEKLVTDRPDY
MUL_1556|M.ulcerans_Agy99 HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCREWVQKTMEKLVTDRPDY
MLBr_01213|M.leprae_Br4923 HFKVVTYLKMGCPLSTEHVPLIMGNNTPYPQCREWVQTTMTKLFSDRPDY
MAV_3209|M.avium_104 HFKVVTYLKMGCPLSTEQVPLIMGNNAPYPQCREWVQRTMTKLVTDRPDY
MAB_2689|M.abscessus_ATCC_1997 NFKITTYLKMGCPLSTEEVPRIAGSNDPYPDCKKWVDEVMSRIIQEHPDY
**:.*********: :. * : *.. **.* .* : .: .:. ::**:
Mvan_3701|M.vanbaalenii_PYR-1 VFTTVTRPRDAQ-PGDVMPDVYLPIWQAFAESGVKVLGMRDTPWLVRNG-
Mflv_2821|M.gilvum_PYR-GCK VFTTVTRPRDTQ-PGDVMPDVYLPIWQAFAESGVKVLGMRDTPWLVRNG-
MSMEG_3187|M.smegmatis_MC2_155 VFTTSTRPWNIK-PGDVMPSTYLGIWETFNENNIPVLAMRDTPWLTRNG-
TH_1799|M.thermoresistible__bu IFTTSTRPWHTR-HGDRVPEGYLGIWEVLNEAGIPVLAMRDTPWLTKDGG
Mb1592c|M.bovis_AF2122/97 VFTTSTRPWNIK-PGDVMPATYVGIWQTFADNNIPVLAMRDTPWLVKDG-
Rv1565c|M.tuberculosis_H37Rv VFTTSTRPWNIK-PGDVMPATYVGIWQTFADNNIPVLAMRDTPWLVKDG-
MMAR_2380|M.marinum_M VFTTSTRPWNIK-PGDVMPATYIGIWQTFADNNIPVLAVRDTPWMVKDG-
MUL_1556|M.ulcerans_Agy99 VFTTSTRPWNIK-PGDVMPATYIGIWQTFADNNIPVLAVRDTPWMVKDG-
MLBr_01213|M.leprae_Br4923 VFTTSTRPWNTK-PGDVMPATYLGIWQALSDNNIPILAMRDTPWLVKNG-
MAV_3209|M.avium_104 VFTTTTRPWNIK-TGDVMPATYIGIWQTLSDNNIPILGMRDTPWLVKNG-
MAB_2689|M.abscessus_ATCC_1997 VFTTTTRPRSALGDGDVMPESYLGIWSAFDEAGIPVLGMRDTPWLIDKDG
:*** *** ** :* *: **..: : .: :*.:*****: ..
Mvan_3701|M.vanbaalenii_PYR-1 EPFFPSDCLAD-GGDATTCGMPRALVLDDVNPAADLTKRFPMMGMIDMSN
Mflv_2821|M.gilvum_PYR-GCK EPFFPRDCLAD-GGDATTCGMPRALVLDDVNPAADLTNRFPLMKVIDMSS
MSMEG_3187|M.smegmatis_MC2_155 KPYFPADCLAD-GGDAVSCGVKRSKVLSDRNPTLDYLDRFPLMKPLDMSD
TH_1799|M.thermoresistible__bu RAKNPVDCLAD-GGDPVHCGMWRYEVLSDENPTLEWVETYPVLKPLDMSD
Mb1592c|M.bovis_AF2122/97 QPFIPADCLAK-GGNPQSCGIARSKVLVDRNPTLDFVARFPLLKPLDMSD
Rv1565c|M.tuberculosis_H37Rv QPFIPADCLAK-GGNPQSCGIARSKVLVDRNPTLDFVARFPLLKPLDMSD
MMAR_2380|M.marinum_M QPFKPADCLAK-GGDAQSCGIKRSELLSDYNTTLDFVDRFPNLKPIDMSD
MUL_1556|M.ulcerans_Agy99 QPFKPADCLAK-GGDAQSCGIKRSELLSDYNTTLDFVDRFPNLKPIDMSD
MLBr_01213|M.leprae_Br4923 QPSNPADCLAK-GGNAVSCGIKRSNVLADRNPTLNFVAQFSLLKPLDMSD
MAV_3209|M.avium_104 QPFDPADCLAKRGSTAQSCAIKRSDVLSERNQTLDFVGQFPQLKVLDMSD
MAB_2689|M.abscessus_ATCC_1997 TTYTAADCISA-GGTADSCAMDRNRALDDVNPTLAIANRFPLLKILDMTR
. . **:: *. . *.: * * : * : :. : :**:
Mvan_3701|M.vanbaalenii_PYR-1 ALCRDDYCRAAEGNILIYRDSHHLSATYVRSMAPELGRRIGALTGWWP--
Mflv_2821|M.gilvum_PYR-GCK AMCRDDYCRAAEGNILIYRDSHHLTATYVRSMAPELGRRIGALTGWWP--
MSMEG_3187|M.smegmatis_MC2_155 AVCREDHCRVVEGNVLLYHDSHHLSATYMRTMTNELGRQMAAATGWW---
TH_1799|M.thermoresistible__bu AVCRPDFCRVVEGNVLVYHDSHHFSSTYMRSMAPELGRQIAAVTGWWVPQ
Mb1592c|M.bovis_AF2122/97 AICRTDTCRAVEGNVLVYRDSHHLTPTYMRTMTSELGRQIAANTDWW---
Rv1565c|M.tuberculosis_H37Rv AICRTDTCRAVEGNVLVYRDSHHLTPTYMRTMTSELGRQIAANTDWW---
MMAR_2380|M.marinum_M AICRPDYCRAVEGNVLIYRDAHHMTPTYARTMAPELGRQIAAITGWW---
MUL_1556|M.ulcerans_Agy99 AICRPDYCRAVEGNVLIYRDAHHMTPTYARTMAPELGRQIAAIIGWW---
MLBr_01213|M.leprae_Br4923 AICRPDICRAVEGNVLIYHGTHHLSPTYVRTLADELGRQIAENTGWW---
MAV_3209|M.avium_104 AICRADMCRPVEGNVLIYHGAHHMSPTYVRTMAPELGRQIAESTGWW---
MAB_2689|M.abscessus_ATCC_1997 AICRPDKCRVVEGNVLVYHDSHHISATYMRTMAKELGRQIALATGWWRPA
*:** * ** .***:*:*:.:**::.** *::: ****::. .**
Mvan_3701|M.vanbaalenii_PYR-1 ----
Mflv_2821|M.gilvum_PYR-GCK ----
MSMEG_3187|M.smegmatis_MC2_155 ----
TH_1799|M.thermoresistible__bu ----
Mb1592c|M.bovis_AF2122/97 ----
Rv1565c|M.tuberculosis_H37Rv ----
MMAR_2380|M.marinum_M ----
MUL_1556|M.ulcerans_Agy99 ----
MLBr_01213|M.leprae_Br4923 ----
MAV_3209|M.avium_104 ----
MAB_2689|M.abscessus_ATCC_1997 QPGQ