For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQTPSPSPAPVAAADPGTSGARTPGSRGFYRYDLDGLRGIAIALVAVFHIWFGRVSGGVDVFLALSGFFF GGKVIRAALNPAVALSPVAEVIRLVRRLVPALVVVLAGCAVLTILVQPETRWETFADQSLASLGYYQNWE LANTASDYLRAGEAVSPLQHIWSMSVQGQFYVSFLVLIAGLAYLFGRPLGARLRTLVLVVLAALTVASFG YAIVAHQQNQAAAYYDSFARGWELLLGALVGALVPRIRWPMWLRTLLATVALGAVLSCGALIDGVQEFPG PWALVPVGAAMLMILVGANLAGDRLPLPNRLLATRPLVELGAIAYSLYLWHWPLLIFWLSYSGHKHAGFV EGTVLLLVSGVLAYLTNRLVEDPLRHRGSARPAPRPAAPTVPWLARVPRPTMALGSAIVLLGVTLTATSF TWRQHVTVLRSAGKELSVLNPQDYPGARALTEHVRVPTLPMRPTVLEVKQDLPASTRDGCISDFVNPAVV NCTYGDVTADRTIALAGGSHAEHWLPALDMLGKLHHFKVVTYLKMGCPLSTEQVPLIMGNNAPYPQCREW VQRTMTKLVTDRPDYVFTTTTRPWNIKTGDVMPATYIGIWQTLSDNNIPILGMRDTPWLVKNGQPFDPAD CLAKRGSTAQSCAIKRSDVLSERNQTLDFVGQFPQLKVLDMSDAICRADMCRPVEGNVLIYHGAHHMSPT YVRTMAPELGRQIAESTGWW
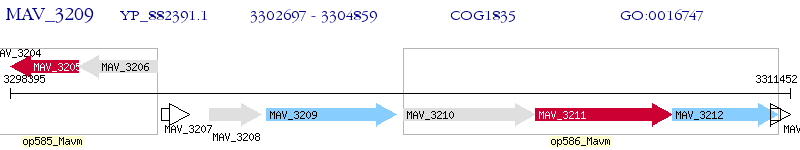
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3209 | - | - | 100% (720) | acyltransferase domain-containing protein |
| M. avium 104 | MAV_4630 | - | 1e-58 | 28.61% (727) | acyltransferase domain-containing protein |
| M. avium 104 | MAV_5201 | - | 7e-32 | 30.27% (413) | putative acyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1592c | - | 0.0 | 74.17% (724) | hypothetical protein Mb1592c |
| M. gilvum PYR-GCK | Mflv_3630 | - | 0.0 | 66.44% (730) | acyltransferase 3 |
| M. tuberculosis H37Rv | Rv1565c | - | 0.0 | 74.17% (724) | hypothetical protein Rv1565c |
| M. leprae Br4923 | MLBr_01213 | - | 0.0 | 76.28% (725) | hypothetical protein MLBr_01213 |
| M. abscessus ATCC 19977 | MAB_2689 | - | 0.0 | 62.69% (721) | acyltransferase |
| M. marinum M | MMAR_2380 | - | 0.0 | 76.14% (725) | hypothetical protein MMAR_2380 |
| M. smegmatis MC2 155 | MSMEG_3187 | - | 0.0 | 69.34% (724) | acyltransferase domain-containing protein |
| M. thermoresistible (build 8) | TH_1799 | - | 0.0 | 67.77% (698) | CONSERVED HYPOTHETICAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1556 | - | 0.0 | 75.86% (725) | hypothetical protein MUL_1556 |
| M. vanbaalenii PYR-1 | Mvan_2787 | - | 0.0 | 69.35% (721) | acyltransferase 3 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3209|M.avium_104 ---------MQTPSPSPA----------PVAAADPGTSGARTPGSRGFYR
MLBr_01213|M.leprae_Br4923 MSSVNWRSIMQTLSPASL----------LVATAKPVPLEGRTAVFTIFYR
Mb1592c|M.bovis_AF2122/97 ---------MLTLSPPRPPA------LTPEPALPPVTMGTRTTG---FYR
Rv1565c|M.tuberculosis_H37Rv ---------MLTLSPPRPPA------LTPEPALPPVTMGTRTTG---FYR
MMAR_2380|M.marinum_M ---------MQTLTPPTP----------PTAAAAPVAMGTRKTG---FYR
MUL_1556|M.ulcerans_Agy99 ---------MQTLTPPTS----------PTAAAAPVAMGTRKTG---FYR
Mflv_3630|M.gilvum_PYR-GCK ---------MMTLAPARPARPAASGLTPPVPAPPASGMGTRSSG---FYR
Mvan_2787|M.vanbaalenii_PYR-1 ---------MMTLAPARPASPTVPGPTRTASVPPSAGMGTRTKG---FYR
MSMEG_3187|M.smegmatis_MC2_155 ----------MTLSPTRPAP-----ISRDTRAANPS-MGTRKSG---FYR
TH_1799|M.thermoresistible__bu ------------------------------------VTGPGTSG---FYR
MAB_2689|M.abscessus_ATCC_1997 ---------MFFVSATKPTK------DPEVSPAAAMDATPKPKGDKAFYR
***
MAV_3209|M.avium_104 YDLDGLRGIAIALVAVFHIWFGRVSGGVDVFLALSGFFFGGKVIRAALNP
MLBr_01213|M.leprae_Br4923 HDLDGLRGIAIALVAIFHVWFGRVSGGVDVLLAMSGFFFGGKILRAALNP
Mb1592c|M.bovis_AF2122/97 HDLDGLRGVAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGKILRAALNP
Rv1565c|M.tuberculosis_H37Rv HDLDGLRGVAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGKILRAALNP
MMAR_2380|M.marinum_M HDLDGLRGIAIALVAIFHVWFGRVSGGVDVFLALSGFFFGGKILRTALNP
MUL_1556|M.ulcerans_Agy99 HDLDGLRGIAIALVAIFHVWFGRVSGGVDVFLALSGFFFGGKILRTALNP
Mflv_3630|M.gilvum_PYR-GCK HDLDGLRGVAIALVAVFHVWFGRVSGGVDVFLVLSGFFFGGRLLRNAITP
Mvan_2787|M.vanbaalenii_PYR-1 HDLDGLRGIAIALVAVFHVWFGRVSGGVDVFLVLSGFFFGGRLLRGALTP
MSMEG_3187|M.smegmatis_MC2_155 HDLDGLRGIAIMMVAVFHIWFGRVSGGVDVFLALSGFFFGGKILRTALDQ
TH_1799|M.thermoresistible__bu YDLDGLRGVAILLVALFHVWFGRVSGGVDVFLALSGFFFGGRLLRAVLAP
MAB_2689|M.abscessus_ATCC_1997 YDLDGLRGIAIFLVAVFHVWFGRVSGGVDVFLTLSGFFYGSKLLRTATTQ
:*******:** :**:**:***********:*.:****:*.:::* .
MAV_3209|M.avium_104 AVALSPVAEVIRLVRRLVPALVVVLAGCAVLTILVQPETRWETFADQSLA
MLBr_01213|M.leprae_Br4923 VPSLSPVAEIIRLVRRLLPALVVVLTGCALLTVVMQPQTRWETFADQSLA
Mb1592c|M.bovis_AF2122/97 DLSLSPIAEVIRLIRRLLPALVVVLAGCALLTIAIQPQTRWEAFANQSLA
Rv1565c|M.tuberculosis_H37Rv DLSLSPIAEVIRLIRRLLPALVVVLAGCALLTIAIQPQTRWEAFANQSLA
MMAR_2380|M.marinum_M SLKLSPITELIRLIRRLLPAMVVVLAACALLTILVQPQTRWEAFADQSLA
MUL_1556|M.ulcerans_Agy99 SLKLSPITELIRLIRRLLPAMVVVLAACALLTILVQPQTRWEAFADQSLA
Mflv_3630|M.gilvum_PYR-GCK GAPLRPVPEVTRLVRRLLPALVVVLAVSAVLTILIQPETRWETFADQSLA
Mvan_2787|M.vanbaalenii_PYR-1 GVALRPVPEVTRLIRRLLPALVVVLAVSAVLTILIQPETRWEAFADQSLA
MSMEG_3187|M.smegmatis_MC2_155 STPLRPLSEVVRLVRRLLPALVVVLAAAAVLTILIQPETRWEAFADQSLA
TH_1799|M.thermoresistible__bu GASLKPLREVTRLVRRLLPALVVVLAASAVLTILIQPTTRWETFADQSLA
MAB_2689|M.abscessus_ATCC_1997 GASLNPVPVVKRLVRRLLPALILVLAACAVLTVLVQPETRWETFAEQSLA
* *: : **:***:**:::**: .*:**: :** ****:**:****
MAV_3209|M.avium_104 SLGYYQNWELANTASDYLRAGEAVSPLQHIWSMSVQGQFYVSFLVLIAGL
MLBr_01213|M.leprae_Br4923 SLGYYQNWELVGTESSYLKAGEAVSPLQHIWSMSVQGQFDIAFLLLVAGC
Mb1592c|M.bovis_AF2122/97 SLGYYQNWELASTVSNYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLVAGC
Rv1565c|M.tuberculosis_H37Rv SLGYYQNWELASTVSNYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLVAGC
MMAR_2380|M.marinum_M SLGYYQNWELANSAADYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLIAGC
MUL_1556|M.ulcerans_Agy99 SLGYYQNWELANSAADYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLIAGC
Mflv_3630|M.gilvum_PYR-GCK SLGYYQNWELARTAADYLRAAETVSPLQHIWSMSVQGQFYIAFLALILLC
Mvan_2787|M.vanbaalenii_PYR-1 SLGYYQNWELARTASDYLRAGETVSPLQHIWSMSVQGQFYIAFLALIMLS
MSMEG_3187|M.smegmatis_MC2_155 SLGYYQNWELANTAADYLRAGETVSPLQHIWSMSVQGQFYIAFLVLIFGF
TH_1799|M.thermoresistible__bu SLGYYQNWELARTASDYLRAGEAVSPLQHIWSMSVQGQFYIAFLVLVMGF
MAB_2689|M.abscessus_ATCC_1997 SLGYYQNWELANTAADYLAASESVSPLQHLWSMSVQGQFYVGFLALVYLL
**********. : :.** *.*:******:********* :.** *:
MAV_3209|M.avium_104 AYLFGRPLG----ARLRTLVLVVLAALTVASFGYAIVAHQQNQAAAYYDS
MLBr_01213|M.leprae_Br4923 AYLFRRPLG----TQLRIMFVVLLGALMIASFIYATFAHQANQTTAYYNS
Mb1592c|M.bovis_AF2122/97 AYLLRRLFRGPRAPYLRTMFVVLLSTLTLASFIYAIVAHHAYQATAYYNT
Rv1565c|M.tuberculosis_H37Rv AYLLRRLFRGPRAPYLRTMFVVLLSTLTLASFIYAIVAHHAYQATAYYNT
MMAR_2380|M.marinum_M TFLFRRLLG----THLRTMFVVLLSALTIASFVYAIIAHQAYQPTAYYNS
MUL_1556|M.ulcerans_Agy99 TFLFRRLLG----THLRTMFVVLLSALTIASFVYAIIAHQAYQPTAYYNS
Mflv_3630|M.gilvum_PYR-GCK AFVFRRVFG----RHARAAFVVLLTGLTVASFVYAVIAHSTDQTTAYYDS
Mvan_2787|M.vanbaalenii_PYR-1 AWLFRRLLG----KRARAGFVVLLGALTIASFVYAIIAHNADQATAYYNS
MSMEG_3187|M.smegmatis_MC2_155 AYLFRRVFG----RHLRTVFIVLLAALTIASFVYAIIAHNTDQATAYYNS
TH_1799|M.thermoresistible__bu AVLFRRVLR----RHLRAAFVVLLTALTVASFVYAIIATNTDQATAYYNS
MAB_2689|M.abscessus_ATCC_1997 AVLLRKPLG----RHIRTVLIVVIAVLSAASFGYAIYAHLDFQSIAYYNT
: :: : : * .:*:: * *** ** * *. ***::
MAV_3209|M.avium_104 FARGWELLLGALVGALVPRIRWPMWLRTLLATVALGAVLSCGALIDGVQE
MLBr_01213|M.leprae_Br4923 FARAWELLLGALVGAAVPYIRWPAWLRTVVATVALAAILSCGALINGVKE
Mb1592c|M.bovis_AF2122/97 FARAWELLAGALVGAVVPHVRWPMWLRTAVATAALAAILSCGALIDGVKE
Rv1565c|M.tuberculosis_H37Rv FARAWELLAGALVGAVVPHVRWPMWLRTAVATAALAAILSCGALIDGVKE
MMAR_2380|M.marinum_M FARAWELLLGALVGALVPYIRWPMWLRTIAATVGLAIVLSCGALINGVKE
MUL_1556|M.ulcerans_Agy99 FTRAWELLLGALVGALVPYIRWPMWLRTIAATVGLAIVLSCGALINGVKE
Mflv_3630|M.gilvum_PYR-GCK FARAWELLLGALVGALVPYVRCPMWLRTVLAVVALAAILSCGALIDGVHE
Mvan_2787|M.vanbaalenii_PYR-1 FARAWELLLGALVGAVVPHVRWPMWLRTLLAVVALAAILSCGALIDGVRE
MSMEG_3187|M.smegmatis_MC2_155 FARAWELLLGALAGALVGFVRWPMWLRTVVSVVSLAAILSCGWFIDGVKE
TH_1799|M.thermoresistible__bu FARAWELLLGALAGSLVPHIRWPKWLRALAATVALAAILSCGVLIDGVNE
MAB_2689|M.abscessus_ATCC_1997 FARAWELLLGVLVGALVAGTRWPRWLRQLLSFVAVVAILSCGALINGVRE
*:*.**** *.*.*: * * * *** : ..: :**** :*:**.*
MAV_3209|M.avium_104 FPGPWALVPVGAAMLMILVGANLAG-----DRLPLPNRLLATRPLVELGA
MLBr_01213|M.leprae_Br4923 FPGPWALVPVGAAMLLILAGANRQSRPGTSASMPLPNRLLATAPLVALGT
Mb1592c|M.bovis_AF2122/97 FPGPWALVPVGATMLMILAGANRQGHPGTRDRLPLPNRLLATAPLVALGA
Rv1565c|M.tuberculosis_H37Rv FPGPWALVPVGATMLMILAGANRQGHPGTRDRLPLPNRLLATAPLVALGA
MMAR_2380|M.marinum_M FPGPWALVPVGAAMLMIMAGANRQAHPSTRDRLPLPNRLLAAGPLVTLGA
MUL_1556|M.ulcerans_Agy99 FPGPWALVPVGAAMLMIMAGANRQAHPSTRDRLPLPNRLLAAGPLVTLGA
Mflv_3630|M.gilvum_PYR-GCK FPGPWALVPVGATLLFILSAANRYADPHTRGRIPAPNRLMATRPFVALGA
Mvan_2787|M.vanbaalenii_PYR-1 FPGPWALVPVGATLLFILSAANRYADPHTRGRIPAPNRLLASKPFVALGA
MSMEG_3187|M.smegmatis_MC2_155 FPGPWALVPVGATILFIFSAANRMSDPRTAGRLPAPNRLLATAPFVSLGS
TH_1799|M.thermoresistible__bu FPGPWALVPVGATILFILSAAN------WQGPLPGPNRLLATTPFVTLGS
MAB_2689|M.abscessus_ATCC_1997 FPGPLALVPVVATLLLILSAANLPAD----ARQPVANRFLATRPLVELGA
**** ***** *::*:*: .** * .**::*: *:* **:
MAV_3209|M.avium_104 IAYSLYLWHWPLLIFWLSYSGHKHAGFVEGTVLLLVSGVLAYLTNRLVED
MLBr_01213|M.leprae_Br4923 IAYTLYLWHWPLLIFWLSYTGHHHANFVEGAALLLVSGLLAYLTTRLIEN
Mb1592c|M.bovis_AF2122/97 MAYSWYLWHWPLLIFWLSYTGHRHANFVEGAAVLLVSGLLAYLTTRLVED
Rv1565c|M.tuberculosis_H37Rv MAYSWYLWHWPLLIFWLSYTGHRHANFVEGAAVLLVSGLLAYLTTRLVED
MMAR_2380|M.marinum_M MAYSLYLWHWPLLIFWLSYTGHRHANFIEGAAILLVSGVLAFLTTRHVED
MUL_1556|M.ulcerans_Agy99 MAYSLYLWHWPLLIFWLSYTGHRHANFIEGAAILLVSGVLAFLTTRHVED
Mflv_3630|M.gilvum_PYR-GCK MAYSLYLWHWPLLIFYLAYTGGTRVNFVEGAVILLVSGILAWLTTRYVEE
Mvan_2787|M.vanbaalenii_PYR-1 MAYSLYLWHWPLLIFYLAHTGNSRVTFVEGAVILLVSGLLAWLTTRYVEE
MSMEG_3187|M.smegmatis_MC2_155 MAYSLYLWHWPLLIFWLSYSGHTAANFVEGAVILLVSGVLAWLTTRYIEE
TH_1799|M.thermoresistible__bu IAYSLYLWHWPLLIFWLAYTGNSGVNFVEGAVILLVSGVLAWLTTKLVED
MAB_2689|M.abscessus_ATCC_1997 LAYSLYLWHWPLLVYWLVYTGEARVTVAEGAAILGISLALAYLTNKYIET
:**: ********:::* ::* . . **:.:* :* **:**.: :*
MAV_3209|M.avium_104 PLRHRGSARPAPRPAAPTVPWLARVPRPTMALGSAIVLLGVTLTATSFTW
MLBr_01213|M.leprae_Br4923 PLRYRTLANTEYPSPARAAAWQLRLHRSTIALGSMVVLLGVALTATSFTW
Mb1592c|M.bovis_AF2122/97 PLRYRAPAGVRSPAAVPPIPWRLRLRRPTIVLGSVVALLGVALTATSFTW
Rv1565c|M.tuberculosis_H37Rv PLRYRAPAGVRSPAAVPPIPWRLRLRRPTIVLGSVVALLGVALTATSFTW
MMAR_2380|M.marinum_M PLRYRAPSGTAAAVKTQAIPWQLRLRRPTIAMGSVVVLLGVTLTATSFTW
MUL_1556|M.ulcerans_Agy99 PLRYRAPSGTAAAVKTQAIPWQLRLRRPTIAMGSVVVLLGVTLTATSFTW
Mflv_3630|M.gilvum_PYR-GCK PLRAGHAPAVAAPKSP--TALR-RLRRPTIVLGSTVALLGVALMATSFTW
Mvan_2787|M.vanbaalenii_PYR-1 PLRSRGTVAATAPVSK--VALRKRLRRPTIVLGSTVALLGVALTATSFTW
MSMEG_3187|M.smegmatis_MC2_155 PLR---TQPGRAPVPA--VPLRARLRRPTIVLGSIVALLGVALTATSFTW
TH_1799|M.thermoresistible__bu PLRYRKPAADVVVPVK--PPLRTRLRRPTMVLGSVVALLGVALTATSFTW
MAB_2689|M.abscessus_ATCC_1997 PLRYPRVSPTSA-------SLWTRLRRPTLALGTSIVLMAVALTATSFTW
*** . *: *.*:.:*: :.*:.*:* ******
MAV_3209|M.avium_104 RQHVTVLRSAGKELSVLNPQDYPGARALTEHVRVPTLPMRPTVLEVKQDL
MLBr_01213|M.leprae_Br4923 RQHVIVLRATGKELSALNAQDYPGARALTAHARVPTLPMRPTVLEIKDDL
Mb1592c|M.bovis_AF2122/97 REHVIVQRAAGKELSGLSSRDYPGARALIDHVRVPKLRMRPTVLEVRHDL
Rv1565c|M.tuberculosis_H37Rv REHVIVQRAAGKELSGLSSRDYPGARALIDHVRVPKLRMRPTVLEVRHDL
MMAR_2380|M.marinum_M REHVIVERASGKELRGLSANDYPGARALTDHVRVPKLRMRPTVFEAKGDF
MUL_1556|M.ulcerans_Agy99 REHVIVERASGKELRGLSANDYPGARALTDHVRVPKLRMRPTVFEAKGDF
Mflv_3630|M.gilvum_PYR-GCK REHLTDLRANGGELATLPLRDYPGARALINDAKVPSVPMRPTVLEAKDDI
Mvan_2787|M.vanbaalenii_PYR-1 REHVTVLRANGKELSVLSLRGYPGALALVDNARVPELPMRPTVLEAKDDI
MSMEG_3187|M.smegmatis_MC2_155 REHVTVQRASGKELSGLSARDYPGARALIDHARVPKLPMRPTVLEAKNDL
TH_1799|M.thermoresistible__bu REHVSLQRASGKELSGLAARDYPGARALTEGARVAKLPMRPSVHEAREDL
MAB_2689|M.abscessus_ATCC_1997 LEHVTVQRSNGKELSGLRPRDYPGAGALLYGDRVPDLPMRPTTLEASDDL
:*: *: * ** * ..**** ** :*. : ***:. * *:
MAV_3209|M.avium_104 PASTRDGCISDFVNPAVVNCTYGDVTADRTIALAGGSHAEHWLPALDMLG
MLBr_01213|M.leprae_Br4923 PASTRDGCISDFVNPAVVNCTYGDASANRTIALAGGSHAEHWLPALDVLG
Mb1592c|M.bovis_AF2122/97 PTSTKDGCISDFVNPAIINCTYGDVDAPRTIALAGGSHAEHWLTALDLLG
Rv1565c|M.tuberculosis_H37Rv PTSTKDGCISDFVNPAIINCTYGDVDAPRTIALAGGSHAEHWLTALDLLG
MMAR_2380|M.marinum_M PASTRDGCISDFVNPALVNCTYGDVDASRTIALAGGSHAEHWLPALDLLG
MUL_1556|M.ulcerans_Agy99 PASTRDGCISDFVNPALVNCTYGDVDASRTIALAGGSHAEHWLPALDLLG
Mflv_3630|M.gilvum_PYR-GCK PESTIDGCISDFDNIGIINCTYGDKSATRTIALAGGSHAEHWITALDLLG
Mvan_2787|M.vanbaalenii_PYR-1 PVSTTEGCISDFDNVDIINCTYGDTAATRTIALAGGSHAEHWITALDLLG
MSMEG_3187|M.smegmatis_MC2_155 PISTTEGCISDFANVGVINCTYGDKNATRTIALAGGSHAEHWITALDLLG
TH_1799|M.thermoresistible__bu PISTIDGCISDFDDVSVIKCTYGDESASRTIALAGGSHAEHWITALDLLG
MAB_2689|M.abscessus_ATCC_1997 PITTEQDCISDFRNRAVITCTYGNPHATRTIALAGGSHAEHWITALDILG
* :* :.***** : ::.****: * **************:.***:**
MAV_3209|M.avium_104 KLHHFKVVTYLKMGCPLSTEQVPLIMGNNAPYPQCREWVQRTMTKLVTDR
MLBr_01213|M.leprae_Br4923 QLHHFKVVTYLKMGCPLSTEHVPLIMGNNTPYPQCREWVQTTMTKLFSDR
Mb1592c|M.bovis_AF2122/97 RMHHFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCHQWVQAAMAKLVADH
Rv1565c|M.tuberculosis_H37Rv RMHHFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCHQWVQAAMAKLVADH
MMAR_2380|M.marinum_M RLHHFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCREWVQKTMEKLVTDR
MUL_1556|M.ulcerans_Agy99 RLHHFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCREWVQKTMEKLVTDR
Mflv_3630|M.gilvum_PYR-GCK RMHHFRVVTYLKMGCPLTTEKVPLVMGNNDPYPKCRQWNDRVLPRIIADK
Mvan_2787|M.vanbaalenii_PYR-1 RAHHFKVVTYLKMGCPLTTEEKPLVMGDNRPYPKCREWNERVMPEIIADK
MSMEG_3187|M.smegmatis_MC2_155 QKHGFKVVTYLKMGCPLTTEEVPLVSGDNRPYPKCHEWNQRVMAKLIADH
TH_1799|M.thermoresistible__bu RKHHFKVVTYLKMGCPLTTDEMPRVSGDGRPYPKCRDWNERVLPELIADR
MAB_2689|M.abscessus_ATCC_1997 RQHNFKITTYLKMGCPLSTEEVPRIAGSNDPYPDCKKWVDEVMSRIIQEH
: * *::.*********:*:. * : *.. ***.*:.* : .: .:. ::
MAV_3209|M.avium_104 PDYVFTTTTRPWNIK-TGDVMPATYIGIWQTLSDNNIPILGMRDTPWLVK
MLBr_01213|M.leprae_Br4923 PDYVFTTSTRPWNTK-PGDVMPATYLGIWQALSDNNIPILAMRDTPWLVK
Mb1592c|M.bovis_AF2122/97 PDYVFTTSTRPWNIK-PGDVMPATYVGIWQTFADNNIPVLAMRDTPWLVK
Rv1565c|M.tuberculosis_H37Rv PDYVFTTSTRPWNIK-PGDVMPATYVGIWQTFADNNIPVLAMRDTPWLVK
MMAR_2380|M.marinum_M PDYVFTTSTRPWNIK-PGDVMPATYIGIWQTFADNNIPVLAVRDTPWMVK
MUL_1556|M.ulcerans_Agy99 PDYVFTTSTRPWNIK-PGDVMPATYIGIWQTFADNNIPVLAVRDTPWMVK
Mflv_3630|M.gilvum_PYR-GCK PDFVFTTSTRPWNIR-PGDVMPSSYLGIWKEFSKHGIPVLAMRDTPWMVD
Mvan_2787|M.vanbaalenii_PYR-1 PDFVFTTSTRPWNIK-PGDVMPSSYVGIWQEFSDNNIPVLAMRDTPWMVR
MSMEG_3187|M.smegmatis_MC2_155 PDYVFTTSTRPWNIK-PGDVMPSTYLGIWETFNENNIPVLAMRDTPWLTR
TH_1799|M.thermoresistible__bu PDYIFTTSTRPWHTR-HGDRVPEGYLGIWEVLNEAGIPVLAMRDTPWLTK
MAB_2689|M.abscessus_ATCC_1997 PDYVFTTTTRPRSALGDGDVMPESYLGIWSAFDEAGIPVLGMRDTPWLID
**::***:*** ** :* *:***. : . .**:*.:*****:
MAV_3209|M.avium_104 -NGQPFDPADCLAKRGSTAQSCAIKRSDVLSERNQTLDFVGQFPQLKVLD
MLBr_01213|M.leprae_Br4923 -NGQPSNPADCLAK-GGNAVSCGIKRSNVLADRNPTLNFVAQFSLLKPLD
Mb1592c|M.bovis_AF2122/97 -DGQPFIPADCLAK-GGNPQSCGIARSKVLVDRNPTLDFVARFPLLKPLD
Rv1565c|M.tuberculosis_H37Rv -DGQPFIPADCLAK-GGNPQSCGIARSKVLVDRNPTLDFVARFPLLKPLD
MMAR_2380|M.marinum_M -DGQPFKPADCLAK-GGDAQSCGIKRSELLSDYNTTLDFVDRFPNLKPID
MUL_1556|M.ulcerans_Agy99 -DGQPFKPADCLAK-GGDAQSCGIKRSELLSDYNTTLDFVDRFPNLKPID
Mflv_3630|M.gilvum_PYR-GCK EDGEPFFPSDCLAD-GGDPVSCGIERSKVLSDRNPTLDYVTRFPLLKPLD
Mvan_2787|M.vanbaalenii_PYR-1 -DGDPFFPADCLAD-GGDAISCGIERSEVLSDHNPTLDYVKRFPMLKPLD
MSMEG_3187|M.smegmatis_MC2_155 -NGKPYFPADCLAD-GGDAVSCGVKRSKVLSDRNPTLDYLDRFPLMKPLD
TH_1799|M.thermoresistible__bu DGGRAKNPVDCLAD-GGDPVHCGMWRYEVLSDENPTLEWVETYPVLKPLD
MAB_2689|M.abscessus_ATCC_1997 KDGTTYTAADCISA-GGTADSCAMDRNRALDDVNPTLAIANRFPLLKILD
.* . . **:: *. . *.: * * : * ** :. :* :*
MAV_3209|M.avium_104 MSDAICRADMCRPVEGNVLIYHGAHHMSPTYVRTMAPELGRQIAESTGWW
MLBr_01213|M.leprae_Br4923 MSDAICRPDICRAVEGNVLIYHGTHHLSPTYVRTLADELGRQIAENTGWW
Mb1592c|M.bovis_AF2122/97 MSDAICRTDTCRAVEGNVLVYRDSHHLTPTYMRTMTSELGRQIAANTDWW
Rv1565c|M.tuberculosis_H37Rv MSDAICRTDTCRAVEGNVLVYRDSHHLTPTYMRTMTSELGRQIAANTDWW
MMAR_2380|M.marinum_M MSDAICRPDYCRAVEGNVLIYRDAHHMTPTYARTMAPELGRQIAAITGWW
MUL_1556|M.ulcerans_Agy99 MSDAICRPDYCRAVEGNVLIYRDAHHMTPTYARTMAPELGRQIAAIIGWW
Mflv_3630|M.gilvum_PYR-GCK MSDAVCREDYCRAVEGNVLMYHDAHHISSTYMRTMTSELGRQIGAATGWW
Mvan_2787|M.vanbaalenii_PYR-1 MSDAVCRADHCRAVEGNVLLYHDAHHLSATYMRTMTDELGRQIGAATGWW
MSMEG_3187|M.smegmatis_MC2_155 MSDAVCREDHCRVVEGNVLLYHDSHHLSATYMRTMTNELGRQMAAATGWW
TH_1799|M.thermoresistible__bu MSDAVCRPDFCRVVEGNVLVYHDSHHFSSTYMRSMAPELGRQIAAVTGWW
MAB_2689|M.abscessus_ATCC_1997 MTRAICRPDKCRVVEGNVLVYHDSHHISATYMRTMAKELGRQIALATGWW
*: *:** * ** ******:*:.:**::.** *::: *****:. .**
MAV_3209|M.avium_104 -------
MLBr_01213|M.leprae_Br4923 -------
Mb1592c|M.bovis_AF2122/97 -------
Rv1565c|M.tuberculosis_H37Rv -------
MMAR_2380|M.marinum_M -------
MUL_1556|M.ulcerans_Agy99 -------
Mflv_3630|M.gilvum_PYR-GCK TS-----
Mvan_2787|M.vanbaalenii_PYR-1 SD-----
MSMEG_3187|M.smegmatis_MC2_155 -------
TH_1799|M.thermoresistible__bu VPQ----
MAB_2689|M.abscessus_ATCC_1997 RPAQPGQ