For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TEPVDTCDLLCLDLPHAESIRATVPDSSTIQSAAAAARGLSDATRLSIAAALAAGDELCVCDMAWVVGLP QGLVSHHLRQLKNASLVSSRRQGKLVMYQLTERGRQLTDSVLASAVALTEKETDRV*
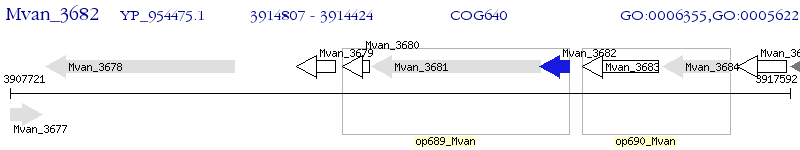
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3682 | - | - | 100% (127) | regulatory protein, ArsR |
| M. vanbaalenii PYR-1 | Mvan_3238 | - | 1e-50 | 76.19% (126) | Fis family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3217 | - | 3e-10 | 36.00% (100) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2675 | - | 1e-09 | 35.37% (82) | ArsR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0907 | - | 2e-55 | 83.59% (128) | regulatory protein, ArsR |
| M. tuberculosis H37Rv | Rv2642 | - | 1e-09 | 35.37% (82) | ArsR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4861c | - | 1e-09 | 38.27% (81) | ArsR family transcriptional regulator |
| M. marinum M | MMAR_2056 | - | 1e-09 | 33.66% (101) | ArsR family transcriptional regulator |
| M. avium 104 | MAV_2037 | - | 3e-07 | 33.33% (78) | transcriptional regulator, ArsR family protein |
| M. smegmatis MC2 155 | MSMEG_4486 | - | 6e-09 | 34.48% (87) | transcriptional regulator, ArsR family protein |
| M. thermoresistible (build 8) | TH_1047 | - | 8e-10 | 34.62% (104) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_3287 | - | 7e-10 | 33.66% (101) | ArsR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2675|M.bovis_AF2122/97 ------------------MSN----------------------LHPLPEV
Rv2642|M.tuberculosis_H37Rv ------------------MSN----------------------LHPLPEV
MMAR_2056|M.marinum_M ------------------MSN----------------------RVEM-DA
MUL_3287|M.ulcerans_Agy99 ------------------MSN----------------------RVEM-DA
TH_1047|M.thermoresistible__bu MFTTRSLDDGFDDYQYRYMSKS--------------------LWESIDGA
MAB_4861c|M.abscessus_ATCC_199 ------------------MPK---------------------ALPVIDVS
Mvan_3682|M.vanbaalenii_PYR-1 ------------------MT-------------------EPVDTCDLLCL
Mflv_0907|M.gilvum_PYR-GCK ------------------MS-------------------EPVDTCDLLCL
MAV_2037|M.avium_104 ----------------MTMSPSPASMPAATDGDMGPEPGLPAHDHDGLAA
MSMEG_4486|M.smegmatis_MC2_155 ------------------MT------------EHG-----EAHDH-----
*.
Mb2675|M.bovis_AF2122/97 ASCVVAPLVREPLNPPAAAEMAARFKALADPVRLQLLSSVASRAGGEACV
Rv2642|M.tuberculosis_H37Rv ASCVVAPLVREPLNPPAAAEMAARFKALADPVRLQLLSSVASRAGGEACV
MMAR_2056|M.marinum_M PCCVAAPLLREPLEASSAVEMAARFKALADPVRLQLLSSVASHAGGEACV
MUL_3287|M.ulcerans_Agy99 PCCVTAPLLREPLEASSAVEMAARFKALADPVRLQLLSSVASHAGGEACV
TH_1047|M.thermoresistible__bu ATCPPAALLREPLSAAEAADVATTLKALADPVRLQLFSAIASHAGGEACV
MAB_4861c|M.abscessus_ATCC_199 APICCPPLSAAPMGEDVAVELALRLKALADPTRLRLLSILLTASDGEVCG
Mvan_3682|M.vanbaalenii_PYR-1 DLPHAESIRATVPDSSTIQSAAAAARGLSDATRLSIAAALAAG--DELCV
Mflv_0907|M.gilvum_PYR-GCK DLPHAEAIRATVPDSATIAAAAAAARGLADATRLSIAAALAAG--DELCV
MAV_2037|M.avium_104 AGHPGPADYPVPPPRDILDAAGELLRALAAPVRIAIVLQLRQ---SHRCV
MSMEG_4486|M.smegmatis_MC2_155 AGTPAPA----LPSRDVLETAGELLRALAAPLRIAIVLQLQQ---SQRCV
. . :.*: . *: : : .. *
Mb2675|M.bovis_AF2122/97 CDISAGVEVSQPTISHHLKVLRDAGLLTSRRRASWVYYAVVPEALTVLSN
Rv2642|M.tuberculosis_H37Rv CDISAGVEVSQPTISHHLKVLRDAGLLTSRRRASWVYYAVVPEALTVLSN
MMAR_2056|M.marinum_M CDISAGVEVGQPTISHHLKVLRDAGLLTSQRRASWVYYAVVPEALNALSR
MUL_3287|M.ulcerans_Agy99 CDISAGVEVGQPTISHHLKVLRDAGLLTSQRRASWVYYAVVPEALNALSR
TH_1047|M.thermoresistible__bu CDISTGLEVSQPTVSHHLKVLREAGLLTSERRGSWVYYAVVPQALASLSA
MAB_4861c|M.abscessus_ATCC_199 CDLASAVGLTEATVSHHLSQLKKAGLAASEKRGLNVFHRANPQAIEALRG
Mvan_3682|M.vanbaalenii_PYR-1 CDMAWVVGLPQGLVSHHLRQLKNASLVSSRRQGKLVMYQLTERGRQLTDS
Mflv_0907|M.gilvum_PYR-GCK CDMAWVVGLPQGLVSHHLRQLKNAFLVSSRRQGKLVMYQLTERGHQLTAA
MAV_2037|M.avium_104 HELVDALGVPQPLVSQHLKILKAAGVVAGERSGREVLYRLAD--HHLAHI
MSMEG_4486|M.smegmatis_MC2_155 HELVDALDVPQPLVSQHLRILKQAGVVSSERAGREVLYRLVD--HHLAHI
:: : : : :*:** *: * : :..: . * :
Mb2675|M.bovis_AF2122/97 LLSVHADAAPALGAPA-
Rv2642|M.tuberculosis_H37Rv LLSVHADAAPALGAPA-
MMAR_2056|M.marinum_M LLSVHAGSATELETSA-
MUL_3287|M.ulcerans_Agy99 LLSVHAGSATELETSA-
TH_1047|M.thermoresistible__bu LLGATGTAP--VGAPR-
MAB_4861c|M.abscessus_ATCC_199 VLMTCC-----------
Mvan_3682|M.vanbaalenii_PYR-1 VLASA-VALTEKETDRV
Mflv_0907|M.gilvum_PYR-GCK VLTGVDTAAAGKDPERV
MAV_2037|M.avium_104 VVDAVAHAGEEPRP---
MSMEG_4486|M.smegmatis_MC2_155 VVDAIAHASEDRR----
::