For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEPVDTCDLLCLDLPHAEAIRATVPDSATIAAAAAAARGLADATRLSIAAALAAGDELCVCDMAWVVGLP QGLVSHHLRQLKNAFLVSSRRQGKLVMYQLTERGHQLTAAVLTGVDTAAAGKDPERV*
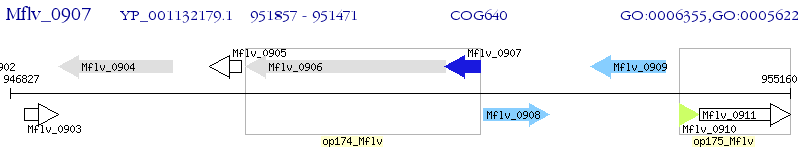
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0907 | - | - | 100% (128) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_2832 | - | 2e-37 | 66.67% (114) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_2718 | - | 2e-10 | 35.58% (104) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2675 | - | 1e-09 | 45.83% (72) | ArsR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2642 | - | 1e-09 | 45.83% (72) | ArsR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4861c | - | 1e-09 | 39.51% (81) | ArsR family transcriptional regulator |
| M. marinum M | MMAR_2056 | - | 8e-09 | 37.33% (75) | ArsR family transcriptional regulator |
| M. avium 104 | MAV_2037 | - | 1e-08 | 36.00% (100) | transcriptional regulator, ArsR family protein |
| M. smegmatis MC2 155 | MSMEG_1173 | - | 1e-09 | 37.89% (95) | transcriptional regulator, ArsR family protein |
| M. thermoresistible (build 8) | TH_1047 | - | 1e-11 | 35.78% (109) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_3287 | - | 6e-09 | 37.33% (75) | ArsR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3682 | - | 2e-55 | 83.59% (128) | regulatory protein, ArsR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0907|M.gilvum_PYR-GCK --------------------MSEPVDTCDLLCLDLPHAEAIRATVPDSAT
Mvan_3682|M.vanbaalenii_PYR-1 --------------------MTEPVDTCDLLCLDLPHAESIRATVPDSST
MSMEG_1173|M.smegmatis_MC2_155 --------------------MSN-QIAATDLAACCPPGALLREPLS-AAE
TH_1047|M.thermoresistible__bu --MFTTRSLDDGFDDYQYRYMSKSLWESIDGAATCPPAALLREPLS-AAE
Mb2675|M.bovis_AF2122/97 --------------------MSN--LHPLPEVASCVVAPLVREPLN-PPA
Rv2642|M.tuberculosis_H37Rv --------------------MSN--LHPLPEVASCVVAPLVREPLN-PPA
MMAR_2056|M.marinum_M --------------------MSN--RVEM-DAPCCVAAPLLREPLE-ASS
MUL_3287|M.ulcerans_Agy99 --------------------MSN--RVEM-DAPCCVTAPLLREPLE-ASS
MAB_4861c|M.abscessus_ATCC_199 --------------------MPK-ALPVIDVSAPICCPPLSAAPMG-EDV
MAV_2037|M.avium_104 MTMSPSPASMPAATDGDMGPEPGLPAHDHDGLAAAGHPGPADYPVPPPRD
. .:
Mflv_0907|M.gilvum_PYR-GCK IAAAA-AAARGLADATRLSIAAALA--AGDELCVCDMAWVVGLPQGLVSH
Mvan_3682|M.vanbaalenii_PYR-1 IQSAA-AAARGLSDATRLSIAAALA--AGDELCVCDMAWVVGLPQGLVSH
MSMEG_1173|M.smegmatis_MC2_155 AADMS-VKLKALADPVRLQLFSAVASREGGEACVCDIAAGVEVSQPTVSH
TH_1047|M.thermoresistible__bu AADVA-TTLKALADPVRLQLFSAIASHAGGEACVCDISTGLEVSQPTVSH
Mb2675|M.bovis_AF2122/97 AAEMA-ARFKALADPVRLQLLSSVASRAGGEACVCDISAGVEVSQPTISH
Rv2642|M.tuberculosis_H37Rv AAEMA-ARFKALADPVRLQLLSSVASRAGGEACVCDISAGVEVSQPTISH
MMAR_2056|M.marinum_M AVEMA-ARFKALADPVRLQLLSSVASHAGGEACVCDISAGVEVGQPTISH
MUL_3287|M.ulcerans_Agy99 AVEMA-ARFKALADPVRLQLLSSVASHAGGEACVCDISAGVEVGQPTISH
MAB_4861c|M.abscessus_ATCC_199 AVELA-LRLKALADPTRLRLLSILLTASDGEVCGCDLASAVGLTEATVSH
MAV_2037|M.avium_104 ILDAAGELLRALAAPVRIAIVLQLR---QSHRCVHELVDALGVPQPLVSQ
: :.*: ..*: : : .. * :: : : : :*:
Mflv_0907|M.gilvum_PYR-GCK HLRQLKNAFLVSSRRQGKLVMYQLTERGHQLTAAVLTGVDTAAAGKDPER
Mvan_3682|M.vanbaalenii_PYR-1 HLRQLKNASLVSSRRQGKLVMYQLTERGRQLTDSVLASA-VALTEKETDR
MSMEG_1173|M.smegmatis_MC2_155 HLKVLRDAGLLTSQRRASWVYYAVVPDALASLAVLLG--ADLRAEVTA--
TH_1047|M.thermoresistible__bu HLKVLREAGLLTSERRGSWVYYAVVPQALASLSALLG--ATGTAPVGAPR
Mb2675|M.bovis_AF2122/97 HLKVLRDAGLLTSRRRASWVYYAVVPEALTVLSNLLSVHADAAPALGAPA
Rv2642|M.tuberculosis_H37Rv HLKVLRDAGLLTSRRRASWVYYAVVPEALTVLSNLLSVHADAAPALGAPA
MMAR_2056|M.marinum_M HLKVLRDAGLLTSQRRASWVYYAVVPEALNALSRLLSVHAGSATELETSA
MUL_3287|M.ulcerans_Agy99 HLKVLRDAGLLTSQRRASWVYYAVVPEALNALSRLLSVHAGSATELETSA
MAB_4861c|M.abscessus_ATCC_199 HLSQLKKAGLAASEKRGLNVFHRANPQAIEALRGVLMTCC----------
MAV_2037|M.avium_104 HLKILKAAGVVAGERSGREVLYRLADHHLAHIVVDAVAHAGEEPRP----
** *: * : :..: . * :
Mflv_0907|M.gilvum_PYR-GCK V
Mvan_3682|M.vanbaalenii_PYR-1 V
MSMEG_1173|M.smegmatis_MC2_155 -
TH_1047|M.thermoresistible__bu -
Mb2675|M.bovis_AF2122/97 -
Rv2642|M.tuberculosis_H37Rv -
MMAR_2056|M.marinum_M -
MUL_3287|M.ulcerans_Agy99 -
MAB_4861c|M.abscessus_ATCC_199 -
MAV_2037|M.avium_104 -