For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSREIDPQLLARLNARRTSRRRFIGGGAAAAAGLALGSSFLAACGSDSGTSSTTSEASGPASGTLRISNW PLYMADGFVAAFQTASGITVDYKEDFNDNEQWFAKVKEPLSRKQDIGADLAVPTSFLAVRLHQLGWLNDI SDEGVPNKKNIRPDLLEASVDPGRKFSAPYMSGLVGLAYNRAATGRDIKTIDDLWDPAFKGRVSLFSDAQ DGLGMIMLSQGNSPENPSMESVQKAVDLVREQNDKGQIRRFTGNDYADDLAAGNVAVAQAYSGDVVQLQA DNPDLQFIVPESGATTFVDTMVIPYTTQNQKAAEAWINYVYDRANYAKLVSYVQYVPVLSDMTEELEKID PAAAANPLINPPADVLAKSKGWAALTDEQTQEYNTAYAAVTGG
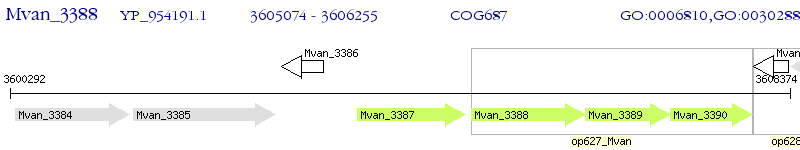
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3388 | - | - | 100% (393) | extracellular solute-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3137 | - | 0.0 | 88.58% (394) | extracellular solute-binding protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02095 | phoS2 | 2e-05 | 31.19% (109) | PstS component of phosphate uptake |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3280 | - | 1e-178 | 77.83% (397) | polyamine-binding lipoprotein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3388|M.vanbaalenii_PYR-1 MSREIDPQLLARLNARRTSRRRFIGGGAAAAAGLALGSSFLAACGS-DSG
Mflv_3137|M.gilvum_PYR-GCK MSRDIDPQLLAQLTARRTSRRRFIGGSAAAAAGLTLGASFLAACGSSDTG
MSMEG_3280|M.smegmatis_MC2_155 MPNNIDPQLFARLTANRTSRRRFLGGGAAAAAALALGPAVLAACGSGGNG
MLBr_02095|M.leprae_Br4923 MKLNQFGAAIGLLATGALLSGCGSDNNAAVGSARTGPSSGQVSCGGKPTL
* : :. * : ...**..:. : .: .:**. .
Mvan_3388|M.vanbaalenii_PYR-1 TSSTTSEAS---GPASGTLRISNWPLYMADGFVAAFQTASGITVDYKEDF
Mflv_3137|M.gilvum_PYR-GCK TSGTTDDG----GPASGTLRISNWPLYMADGFVAAFQTASGITVDYKEDY
MSMEG_3280|M.smegmatis_MC2_155 GSEATSAAPDDGSPASGTLRISNWPLYMADGFVAAFQTASGLTVDYKEDF
MLBr_02095|M.leprae_Br4923 KASGSTAQAN-----AMTRFVNAFERSCPGQTLNYTANGSGAGVS---EF
: : : * :. : .. : ..** *. ::
Mvan_3388|M.vanbaalenii_PYR-1 NDNEQWFAKVKEPLSRKQDIGADLAVPTSFLAVRLHQLGWLNDISDEGVP
Mflv_3137|M.gilvum_PYR-GCK NDNEQWFAKVKEPLSRKQDIGADLVVPTEFMAIRLHQLGWLNDISDEGVP
MSMEG_3280|M.smegmatis_MC2_155 NDNEQWFAKVKEPLSRKQDIGADLVVPTEFMAARLMGLNWLNEINESRVP
MLBr_02095|M.leprae_Br4923 NGNQTDFGGSDSPLSRKEYAAAEQRCGSQAWNLPVVFGPIAITYNVNGLS
*.*: *. ..*****: .*: :. : . . :.
Mvan_3388|M.vanbaalenii_PYR-1 NKKNIRPDLLEASVDPGRKFSAPYMSGL-VGLAYNRAATGRDIKTIDDLW
Mflv_3137|M.gilvum_PYR-GCK NKKNLRPDLMEASADPGRKFSAPYMSGL-VGLAYNRAATGRDISSIDDLW
MSMEG_3280|M.smegmatis_MC2_155 NKKNLREDLLDSKVDPGRKYTAPYMTGM-VGIAYNSAATGREITKIDDLW
MLBr_02095|M.leprae_Br4923 SLNLDGPTTAKIFNGSIASWNDPAIQALNTGVALPAEPIHVVFRNDESGT
. : . .. .:. * : .: .*:* . : . :.
Mvan_3388|M.vanbaalenii_PYR-1 DPAFKGRVSLFSDAQDGLGMIMLSQGNSPENPSMESVQKAVDLVREQNDK
Mflv_3137|M.gilvum_PYR-GCK DPAFKGRVSLLSDTQDGLGMIMLSQGNSPENPSTESVQRAVDLVREQNDR
MSMEG_3280|M.smegmatis_MC2_155 DPAFKGRVSLLSDVQDGLGMIMQWQGNSVEDPTTEGVQKAADFVREQKDK
MLBr_02095|M.leprae_Br4923 TDNFQRYLDVASNGEWGKGIGKTFKGGVGEG--------------AKGND
*: :.: *: : * *: :*. *. : :
Mvan_3388|M.vanbaalenii_PYR-1 GQIRRFTGNDYADDLAAGNVAVAQAYSGDVVQLQADNPDLQFIVPESGAT
Mflv_3137|M.gilvum_PYR-GCK GQIRRFTGNDYADDLAAGNIAVAQAYSGDVVQLQADNPDLQFIVPQSGGT
MSMEG_3280|M.smegmatis_MC2_155 GQIRRFTGNDYADDLASGNVAVAQAYSGDVVQLQADNPDLKFIVPESGGD
MLBr_02095|M.leprae_Br4923 GTSAAVKSTEGSITYNEWSFASARKLNTAKIATSADPEPIAISVDSVGKT
* ....: : ..* *: . : .** : : * . *
Mvan_3388|M.vanbaalenii_PYR-1 TFVDTMVIPYTTQNQKAAEAWINYVYDRANYAKLVSYVQYVPVLSDMTEE
Mflv_3137|M.gilvum_PYR-GCK TFLDTMVIPYTTQNQKAAEAWIDYVYDRANYAKLVAFTQFVPVLSEMTEE
MSMEG_3280|M.smegmatis_MC2_155 WFIDTMVIPYTTQNQKAAEAWIDYVYDRPNYAKLVAFTQFVPVLSDMTDE
MLBr_02095|M.leprae_Br4923 ISGATIIG----EGNDLVLDTVSFYKPAQPGSYPIVLATYEIVCSKYPDA
*:: :.:. . :.: : : . : * *. .:
Mvan_3388|M.vanbaalenii_PYR-1 LEKIDPAAAANPLINPPADVLAKSKGWAALTDEQTQEYNTAYAAVTGG
Mflv_3137|M.gilvum_PYR-GCK LEKVDPAAASNPLINPPADVLERCKSWAALTDEQTQEFNTAYAAVTGG
MSMEG_3280|M.smegmatis_MC2_155 LAKIDQALADNPLINPPAEMAANLKSWAALTDEQTQEFNTIYAEVTGG
MLBr_02095|M.leprae_Br4923 QVGRAVKAFLQSTIGGGQNGLGD-NGYVPIPDSFKSRLSTAANAIA--
:. *. : :.:..:.*. ... .* ::