For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TLIRTTLSAAVVACGLAAGMLSSGAPAGAQPAPPPAPVQGEVPPAWAPPKPAETWAGHPVVWYSGWGGRW GVWINGSFISLTSNPVTGGG*
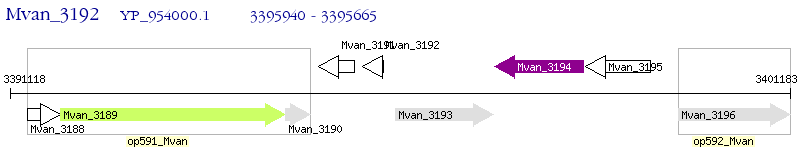
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3192 | - | - | 100% (91) | hypothetical protein Mvan_3192 |
| M. vanbaalenii PYR-1 | Mvan_2955 | - | 1e-15 | 51.25% (80) | hypothetical protein Mvan_2955 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3438 | - | 2e-37 | 78.02% (91) | hypothetical protein Mflv_3438 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1439c | - | 1e-05 | 43.64% (55) | hypothetical protein MAB_1439c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1245 | - | 1e-06 | 40.00% (60) | secreted protein antigen |
| M. smegmatis MC2 155 | MSMEG_3454 | - | 1e-15 | 51.85% (81) | hypothetical protein MSMEG_3454 |
| M. thermoresistible (build 8) | TH_3438 | - | 4e-06 | 40.00% (60) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3192|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3438|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3454|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1439c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1245|M.avium_104 --------------------------------------------------
TH_3438|M.thermoresistible__bu VTNWTGGGDFGGPNPIGGDPSGGQPGGGWGAPPPPPTQWTPEQPPQWGNP
Mvan_3192|M.vanbaalenii_PYR-1 ---------------------------------------MTLIR----TT
Mflv_3438|M.gilvum_PYR-GCK ---------------------------------------MALLKTWMKTA
MSMEG_3454|M.smegmatis_MC2_155 ---------------------------------------MSDPNTEVTYM
MAB_1439c|M.abscessus_ATCC_199 ---------------------------------------MNLKSLVAQGA
MAV_1245|M.avium_104 ----------------------------------------MTTEKRTAGW
TH_3438|M.thermoresistible__bu QQPGFPPPGPQQFGPAQPGWGGQWGQPPPPDGPKRNRKALLVTLGAGAAV
Mvan_3192|M.vanbaalenii_PYR-1 LSAAVVACGLAAG---MLSSG-APAGAQPAPPPAPVQ-------------
Mflv_3438|M.gilvum_PYR-GCK LPAAVMAGGLAAA---VLGSGAAPAFAQPAPPPAPV--------------
MSMEG_3454|M.smegmatis_MC2_155 ILIRAALAGVAAAGV-VLGGAMGLATASADPVPPPP--------------
MAB_1439c|M.abscessus_ATCC_199 AVGALAMGTLGMGAVPAFAQPVGHPAYTQDHPNRPHGDDDDDWGGHDGR-
MAV_1245|M.avium_104 AASVVAGGALAMSVVGLAGGPVAAAAPAPAPTGHWCPGDPWNPSWG----
TH_3438|M.thermoresistible__bu LVVLVVVGVIALSGLGGSGDGGSSRSAGAAVQGYLEALARGDAEAALSYG
:. . . .
Mvan_3192|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3438|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3454|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1439c|M.abscessus_ATCC_199 -----------HNNGNWRGDDSRRDRDRDWRDDGRWQDDRREWRDNPWHY
MAV_1245|M.avium_104 ---------------------------------NVLDWDWHQCHDWQHPG
TH_3438|M.thermoresistible__bu AGEPASKELLTDEVLKSQIEKMPISNIRILSDDSDNGVGFGKVHATVNFG
Mvan_3192|M.vanbaalenii_PYR-1 GEVPP-AWAPPKPAETWAG----------HPVVWYSG------WGG----
Mflv_3438|M.gilvum_PYR-GCK GEQPP-AWAPPKPAEIWNG----------HPVVWNSA------WGG----
MSMEG_3454|M.smegmatis_MC2_155 GPPVP-PNAPKKPAETWQG----------HPMVWAQLP-----AGG----
MAB_1439c|M.abscessus_ATCC_199 DQRPPWGWAPP-PRAEWRGP---------LPDRWGPPPQAFDYWGHRVQP
MAV_1245|M.avium_104 GPTGPAGWGPWGPPPAWAPP---------PPPQPAWAPGAQMMWNPTGAG
TH_3438|M.thermoresistible__bu DQVSDVTFLMKKSGNVWKLDSAAVRLDLSTQASSNRAMSTLTVFGKTVGD
. . * .
Mvan_3192|M.vanbaalenii_PYR-1 ----------RWGVWIN-----GSFISLTSNPVTGGG-------------
Mflv_3438|M.gilvum_PYR-GCK ----------RWGAWIN-----GGFISLTSNPVTGGG-------------
MSMEG_3454|M.smegmatis_MC2_155 ----------HWGVWIN-----GDFLPFS---------------------
MAB_1439c|M.abscessus_ATCC_199 VWDDGFR---TWGFWLF-----GIWIPLVSVG------------------
MAV_1245|M.avium_104 ----------NWGIWNN-----GVWTPL----------------------
TH_3438|M.thermoresistible__bu SVLYLFPGYIEWGTNNKNVTAQGQPLLLDALTGYAGGSAPKIVLSDSARK
** * :
Mvan_3192|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3438|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3454|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1439c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1245|M.avium_104 --------------------------------------------------
TH_3438|M.thermoresistible__bu SITSAVRSYLDECARSKSLSPEGCPQRAFEYGAVDGTVTWHAPGTDNLQI
Mvan_3192|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3438|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3454|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1439c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_1245|M.avium_104 --------------------------------------------------
TH_3438|M.thermoresistible__bu SFDGYSMEARVTLNGQFPYTVQGRDGAPINGSDHFMLYGRADLNQTPPVL
Mvan_3192|M.vanbaalenii_PYR-1 ---
Mflv_3438|M.gilvum_PYR-GCK ---
MSMEG_3454|M.smegmatis_MC2_155 ---
MAB_1439c|M.abscessus_ATCC_199 ---
MAV_1245|M.avium_104 ---
TH_3438|M.thermoresistible__bu KWN