For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SYVDPPQFVSKMIDAGENKVFMSARDTVIRAYMAGAILALAAAFAVTITVQTGNALLGAVLFPVGFCLLY LMGFDLLTGVFTLVPLALLDKRRGVTVGSMLRNWGLVFVGNFLGALTVAVMIAVTLTYGFTVDPNEVGQR LGEIGSSRTVGYAEHGAAGMLTLFVRGVLCNWMVSTGVVAAMMSTSVSGKVITMWMPIMLFFYMGFEHSV VNMFLFPSGLMLGGDFSIADYLIWNEIPTVVGNLVGGLAFVGLTLYATHARTGGEWNRPAGSEDFAIPVK ANA*
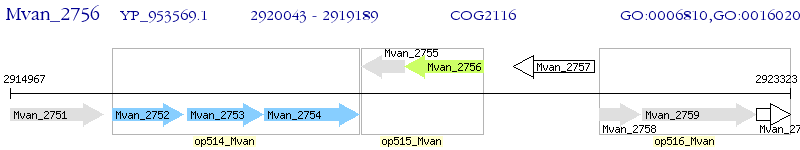
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2756 | - | - | 100% (284) | formate/nitrite transporter |
| M. vanbaalenii PYR-1 | Mvan_4123 | - | 1e-10 | 24.08% (245) | formate/nitrate transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1996 | - | 1e-132 | 87.12% (264) | formate/nitrite transporter |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2490 | - | 2e-09 | 26.12% (245) | formate/nitrite transporter superfamily protein |
| M. smegmatis MC2 155 | MSMEG_5360 | - | 1e-132 | 84.36% (275) | formate/nitrate transporter |
| M. thermoresistible (build 8) | TH_1321 | - | 4e-10 | 23.36% (244) | hypothetical protein MSMEG_4318 |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1996|M.gilvum_PYR-GCK --------MFRIGNTLRELFREHPAPERFHMSYVSPSE--FVGKMIDAGE
MSMEG_5360|M.smegmatis_MC2_155 ------------------------------MSYVSPSD--FVGKMIDAGE
Mvan_2756|M.vanbaalenii_PYR-1 ------------------------------MSYVDPPQ--FVSKMIDAGE
MAV_2490|M.avium_104 MTINASAPQRQGAVAVVDTVTALPAAVAGDGGTADPFEPLTVGAMVDRVS
TH_1321|M.thermoresistible__bu ------VHFAHARGYPTAVSDTSQRDLGDSDSPIEDALEEAFRRMVDEGT
. . . *:*
Mflv_1996|M.gilvum_PYR-GCK SKALMSTRD---TLIRAFMAGAILALAAAFAVTITVQTGN----PLLGAV
MSMEG_5360|M.smegmatis_MC2_155 AKAHMSTRD---TLIRAYMAGAILAIAAAFAVTITVQTGN----ALVGAI
Mvan_2756|M.vanbaalenii_PYR-1 NKVFMSARD---TVIRAYMAGAILALAAAFAVTITVQTGN----ALLGAV
MAV_2490|M.avium_104 AMAVEKAAHPWAFLMRSLVGGAMVAFGVLLALVVSTGVKTPGVASLLMGL
TH_1321|M.thermoresistible__bu QRLHRRWRE---VLVTGFFGGTEIAVGVLAYLAVLHVTHN----HLLAGL
. :: . ..*: :*... :.: . . *: .:
Mflv_1996|M.gilvum_PYR-GCK LFPVGFCLLYLLGFDLLTGVFTLVPLAMLDKRRGVTFCSMMRNWGLVFVG
MSMEG_5360|M.smegmatis_MC2_155 LFPVGFCLLYLLGFDLLTGVFTLVPLALLDKRRGVTVRTVLRNWGWVFLG
Mvan_2756|M.vanbaalenii_PYR-1 LFPVGFCLLYLMGFDLLTGVFTLVPLALLDKRRGVTVGSMLRNWGLVFVG
MAV_2490|M.avium_104 AFGMSFVLILVSGMSLITADMAAGFLAVLQR--ALSIRSYVVLVAVGLVG
TH_1321|M.thermoresistible__bu AFSIGFLALLLGRSELFTEGFLIPVTTVAAKR--ASVAQLMKLWGGTLAA
* :.* : : .*:* : :: : :. : . : .
Mflv_1996|M.gilvum_PYR-GCK NLGGAIVVALMMAVVFTFGFSVDPNEVGQRLGEIGHSRTVGYAEHGAAGM
MSMEG_5360|M.smegmatis_MC2_155 NFAGALTVAFMMAIVFTYGFSVDPNEVGQRIGEIGHSRTVGYAEHGAAGM
Mvan_2756|M.vanbaalenii_PYR-1 NFLGALTVAVMIAVTLTYGFTVDPNEVGQRLGEIGSSRTVGYAEHGAAGM
MAV_2490|M.avium_104 NIVGALVFVTVCAAAG-----------GPYLGAFADRAATVGTQKAGQPF
TH_1321|M.thermoresistible__bu NLVGGWVLMWLIMTAF------------PRLHAQTTESAAHYATAPLS--
*: *. .. : . : :. :
Mflv_1996|M.gilvum_PYR-GCK LTLFIRAVLCNWMVSTGVVAAMMSTSVSGKVIAMWMPIMLFFYMGFEHSI
MSMEG_5360|M.smegmatis_MC2_155 LTLFIRGVLCNWMVSTGVVAAMMSTSVSGKVIGMWMPVMLFFYMGFEHSV
Mvan_2756|M.vanbaalenii_PYR-1 LTLFVRGVLCNWMVSTGVVAAMMSTSVSGKVITMWMPIMLFFYMGFEHSV
MAV_2490|M.avium_104 WTALLLAVLCTWFLQTSMCMFFKARSDVARMALAFYGPFAFVIGGTQHVI
TH_1321|M.thermoresistible__bu AETMCLALLGGMVITLMTRMQHGTDAMIGKIAAAVAGGFLLAGLQMFHSI
: .:* .: : : .:: : : * :
Mflv_1996|M.gilvum_PYR-GCK VNMFLFPSGLMLGGDFSIADY-----------LIWNEIPTVLGNLVGGLT
MSMEG_5360|M.smegmatis_MC2_155 VNMFLFPSGLMLGGDFSIADY-----------FIWNEIPTAVGNLVGGLT
Mvan_2756|M.vanbaalenii_PYR-1 VNMFLFPSGLMLGGDFSIADY-----------LIWNEIPTVVGNLVGGLA
MAV_2490|M.avium_104 ANVGFVGLPLLLNLFHPIAARGDIGWGFGDHGLLTNIGVTTVGNLIGGTV
TH_1321|M.thermoresistible__bu LDSLLIFGALIAG-DAPFGYLD----------WLRWFGYTVVGNVAGGLG
: :. *: . .:. : *.:**: **
Mflv_1996|M.gilvum_PYR-GCK FVGLTLFATHARTGETRLVELPTEDRSLVGAAGRVLR
MSMEG_5360|M.smegmatis_MC2_155 FVGLTLFATHARTAAPR-FSIPAVAEPHSAEARKISA
Mvan_2756|M.vanbaalenii_PYR-1 FVGLTLYATHARTGGEWNRPAGSEDFAIPVKANA---
MAV_2490|M.avium_104 FVALPFWIIAHLQRRRLLSTGALRPDG----------
TH_1321|M.thermoresistible__bu LVTMLRLLRSKERLQEERRDAEAE-------------
:* :