For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VASSSTVHPARYLALFLAMLIGAYLLVFLTGDKKPDPKLGIDLQGGTRVTLTARTPDGSPPSRESLDQAK QIIGARVDGLGVSGSEVIIDGQNLVITVPGDDTSEARSLGQTARLYIRPVVHVIPSAAAAPPEQAGPQGV PPGGVPGVPPGAVPGLPPGAVPPGAPPGAVPGLPPAAVPPEAPPASDTPAPQPRPFPQQPAPTPPPGPSP APGPTPPPAPGTAPPADQDVPLATRIQDEKRLRQSTEQAIQILALQFQATRCGQEDVLAGNDDPNLPLVT CSQDGNEVYLLDKSIISGEQIANASSGLNQQAGEYVVDVEFKSDAAKVWADFTAANVGTQTAFVLDSQVV SAPVIQEAIPGGRTQITGQFTQTTARELANVLKYGSLPLSFESSEAETVSATLGLSSLRAGLIAGVVGLV AVLLYSLLYYRVLGVLIALSLTASGAMVFAILVLLGRYISYTLDLAGIAGLIIGIGMTADSFVVFFERIK DEIREGRSFRSAVPRGWARARKTIVSGNAVTLLAAAVLYFLAVGQVKGFAFTLGLTTILDIVVVFLVTWP LVYMASKSTLWAKPSLNGLGAVQQIARERRAAASAAGRG
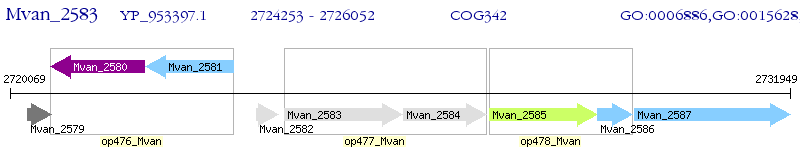
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2583 | secD | - | 100% (599) | preprotein translocase subunit SecD |
| M. vanbaalenii PYR-1 | Mvan_0626 | - | 5e-14 | 39.42% (104) | hypothetical protein Mvan_0626 |
| M. vanbaalenii PYR-1 | Mvan_2314 | infB | 2e-12 | 41.38% (116) | translation initiation factor IF-2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2618c | secD | 0.0 | 71.05% (601) | preprotein translocase subunit SecD |
| M. gilvum PYR-GCK | Mflv_3816 | secD | 0.0 | 79.97% (619) | preprotein translocase subunit SecD |
| M. tuberculosis H37Rv | Rv2587c | secD | 0.0 | 71.21% (601) | preprotein translocase subunit SecD |
| M. leprae Br4923 | MLBr_00487 | secD | 0.0 | 66.89% (607) | preprotein translocase subunit SecD |
| M. abscessus ATCC 19977 | MAB_2880c | secD | 0.0 | 68.81% (606) | preprotein translocase subunit SecD |
| M. marinum M | MMAR_2120 | secD | 0.0 | 70.57% (615) | protein-export membrane protein SecD |
| M. avium 104 | MAV_3468 | secD | 0.0 | 70.75% (612) | preprotein translocase subunit SecD |
| M. smegmatis MC2 155 | MSMEG_2961 | secD | 0.0 | 73.68% (623) | preprotein translocase subunit SecD |
| M. thermoresistible (build 8) | TH_1363 | secD | 1e-154 | 83.23% (334) | PROBABLE PROTEIN-EXPORT MEMBRANE PROTEIN SECD |
| M. ulcerans Agy99 | MUL_1725 | secD | 0.0 | 70.57% (615) | preprotein translocase subunit SecD |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2618c|M.bovis_AF2122/97 --------------------------MASSSAPVHPARYLSVFLVMLIGI
Rv2587c|M.tuberculosis_H37Rv --------------------------MASSSAPVHPARYLSVFLVMLIGI
MMAR_2120|M.marinum_M --------------------------MASSSAPVHPARYLSVFLVLLIGV
MUL_1725|M.ulcerans_Agy99 --------------------------MASSSAPVHPARYLSVFLVLLIGV
MLBr_00487|M.leprae_Br4923 MSCVKGLVMRHVGSVGKNRFRGDTRNVASSSAPVHFGRYLSMFLVLFIGV
MAV_3468|M.avium_104 --------------------------MASSSVPVHPARYLSVFLLLLVGV
Mvan_2583|M.vanbaalenii_PYR-1 --------------------------MASSST-VHPARYLALFLAMLIGA
Mflv_3816|M.gilvum_PYR-GCK --------------------------MASSST-VHPARYLALFLVLLIGA
TH_1363|M.thermoresistible__bu --------------------------------------------------
MSMEG_2961|M.smegmatis_MC2_155 --------------------------MASSSAPVHPARYLTLFLVLLVGV
MAB_2880c|M.abscessus_ATCC_199 --------------------------MASSSAPVHPMRYLAAFLVLLIGA
Mb2618c|M.bovis_AF2122/97 YLLVFFTGDKHTAPKLGIDLQGGTRVTLTARTPDGSAPSREALAQAQQII
Rv2587c|M.tuberculosis_H37Rv YLLVFFTGDKHTAPKLGIDLQGGTRVTLTARTPDGSAPSREALAQAQQII
MMAR_2120|M.marinum_M YLLVFLTGDKKAAPKLGIDLQGGTRVTLTARTPDGSAPSREALAQAQQII
MUL_1725|M.ulcerans_Agy99 YLLVFLTGDKKAAPKLGIDLQGGTRVTLTARTPDGSAPSREALAQAQQII
MLBr_00487|M.leprae_Br4923 YLLAFLTGDKRVVPRLGIDLQGGTRVTLTARTPDGSRPSREALSQAQQII
MAV_3468|M.avium_104 YLLVFLTGDKRAAPKLGIDLQGGTRVTLTARTPDGSAPSREALAQAQQII
Mvan_2583|M.vanbaalenii_PYR-1 YLLVFLTGDKKPDPKLGIDLQGGTRVTLTARTPDGSPPSRESLDQAKQII
Mflv_3816|M.gilvum_PYR-GCK FLLVFFTGDKKPDPKLGIDLQGGTRVTLTARTPDGSEPSRDALLQAQQII
TH_1363|M.thermoresistible__bu --------------------------------------------------
MSMEG_2961|M.smegmatis_MC2_155 YLLVFLTGDKKPEPKLGIDLQGGTRVTLTARTPDGSAPTRDALNQAQQII
MAB_2880c|M.abscessus_ATCC_199 YLLVFLTGDKQPKPKLGIDLQGGTRVTLTARTPDGSKPTKEALTQAQQII
Mb2618c|M.bovis_AF2122/97 SARVNGLGVSGSEVVVDGDNLVITVPGNDGSEARNLGQTARLYIRPVLNS
Rv2587c|M.tuberculosis_H37Rv SARVNGLGVSGSEVVVDGDNLVITVPGNDGSEARNLGQTARLYIRPVLNS
MMAR_2120|M.marinum_M SARVNGLGVSGSEVVVDGDNLVITVPGNDGNEARNLGQTARLYIRPVMNS
MUL_1725|M.ulcerans_Agy99 SARVNGLGVSGSEVVVDGDNLVITVPGNDGNEARNLGQTARLYIRPVMNS
MLBr_00487|M.leprae_Br4923 SSRVNGLGVSGSEVVVDGDNLVITVPGNDGNEARNLGQTARLYIRPVLNS
MAV_3468|M.avium_104 SARVNGLGVSGSEVVVDGDNLIITVPGNDGNEARNLGQTARLYIRPVLNS
Mvan_2583|M.vanbaalenii_PYR-1 GARVDGLGVSGSEVIIDGQNLVITVPGDDTSEARSLGQTARLYIRPVVHV
Mflv_3816|M.gilvum_PYR-GCK SARVDGLGVSGSEVIIDGNNLVITVPGDDTSEARSLGQTARLYIRPVVHA
TH_1363|M.thermoresistible__bu --------------------------------------------------
MSMEG_2961|M.smegmatis_MC2_155 SARVDGLGVSGSEVVIDGDNLVITVPGNDGNEARNLGQTARLYIRPVIHA
MAB_2880c|M.abscessus_ATCC_199 NARVNGLGVSGSEVIIDGDNLVITVPGNDGSEARNLGQTAKLYIRPVVQA
Mb2618c|M.bovis_AF2122/97 MPAQPAAEEPQ-----------PAPSAEPQ--------------------
Rv2587c|M.tuberculosis_H37Rv MPAQPAAEEPQ-----------PAPSAEPQ--------------------
MMAR_2120|M.marinum_M MPAQPAAQEPQQ-EPQQPGGTAPAPGGPAP--------------------
MUL_1725|M.ulcerans_Agy99 MPAQPAAQEPQQ-EPQQPGGTAPAPGGPAP--------------------
MLBr_00487|M.leprae_Br4923 LPVQ---------------------RGQDP--------------------
MAV_3468|M.avium_104 MPAQSGEPKPAPRQPAQPAPGQPAPGQPAPGQP-----------------
Mvan_2583|M.vanbaalenii_PYR-1 IPS-AAAAP---PEQAGPQGVP--PGGVPGVPPGAVPGLP-PGAVP----
Mflv_3816|M.gilvum_PYR-GCK IPAGEGAAPGGAPEGMPPGAIPGMPGGVPGMPPGAIPGMP-PGAIPGMPG
TH_1363|M.thermoresistible__bu --------------------------------------------------
MSMEG_2961|M.smegmatis_MC2_155 MAAQPPTEDGQPPAGAP--GAPGAPGGAPGAP--GLPGLPGLSGLPGMPG
MAB_2880c|M.abscessus_ATCC_199 VPAQPKENP--------------EPG--AGAP------------------
Mb2618c|M.bovis_AF2122/97 ---PPGQP---AAPPPAQ--SGAPASPQ------PGAQPRPYPQDP----
Rv2587c|M.tuberculosis_H37Rv ---PPGQP---AAPPPAQ--SGAPASPQ------PGAQPRPYPQDP----
MMAR_2120|M.marinum_M ---NAGQPPESGAPASPQ--SGAPAAPQSGTPGRPGAQPRPYPQDPPPSP
MUL_1725|M.ulcerans_Agy99 ---NAGQPPESSAPASPQ--SGAPAAPQSGTPGRPGAQPRPYPQDPPPSP
MLBr_00487|M.leprae_Br4923 ---KPGSP-----AGTLP--NFAPMPPD-----HPGAQPRPYLQDP----
MAV_3468|M.avium_104 ---APGQPAPGQPAPGQP--APAPAQPG-----AAPAQPRPYPQDPPPSP
Mvan_2583|M.vanbaalenii_PYR-1 ----PGAPPGAVPGLPPA--AVP--PEAPPASDTPAPQPRPFPQQP----
Mflv_3816|M.gilvum_PYR-GCK DA-MPGMPEGAMP-LPPG--ALQGGPGGPGEPVAPAPQPRPFPQQP----
TH_1363|M.thermoresistible__bu --------------------------------------------------
MSMEG_2961|M.smegmatis_MC2_155 APGAPGAPQEAPPGMPVPGEAPGGEAPAPPAGEQPAPQPRPYPLEP----
MAB_2880c|M.abscessus_ATCC_199 ---QQGQPAPAKEATPPAPGAGKSTPAAPP---SPKPQPRPYPAQT----
Mb2618c|M.bovis_AF2122/97 --APSPNPTSPASPPPA--------PPAEAPAT-DPRKDLAERIAQEKKL
Rv2587c|M.tuberculosis_H37Rv --APSPNPTSPASPPPA--------PPAEAPAT-DPRKDLAERIAQEKKL
MMAR_2120|M.marinum_M APAPSPAPTGSPAPPATGAQQPADLPPGEEPALPDPRKDLAERIAQEKKW
MUL_1725|M.ulcerans_Agy99 APAPSPAPTGSPAPPATGAQQPADLPPGEEPALPDPRKDLAERIAQEKKW
MLBr_00487|M.leprae_Br4923 --TSSPSSDPMPSPVPTGAALPGEVPSVEQPAPPDPRKDLAERIAEEKKW
MAV_3468|M.avium_104 SPAPAPAPAPGPAPAPAPAPAPGEAPPGEQPAPPDPRKDLAERIAEEKKW
Mvan_2583|M.vanbaalenii_PYR-1 --APTPPPGPSPAPGPT-PPP----APGTAPPADQD-VPLATRIQDEKRL
Mflv_3816|M.gilvum_PYR-GCK --APSPAPEPPADPAAP-IPPGTPAGPGGMPADDEN-LSLADQIKQEKLR
TH_1363|M.thermoresistible__bu --------------------------------------------------
MSMEG_2961|M.smegmatis_MC2_155 --APTPG-EPTPAPAPE-PAPGENPPAAPAPGADGDKADLAQRIATAKQW
MAB_2880c|M.abscessus_ATCC_199 --TTPPPPPPATQPATP-NATTAPAPPADLPPKPGSGEERKQLIDFEKQI
Mb2618c|M.bovis_AF2122/97 RQSTNQYMQMVALQFQATRCES-DDILAGNDDPKLPLVTCSTD---HKTA
Rv2587c|M.tuberculosis_H37Rv RQSTNQYMQMVALQFQATRCES-DDILAGNDDPKLPLVTCSTD---HKTA
MMAR_2120|M.marinum_M RQSTNQYIQMVALQFEATQCGK-DDILAGNDDPKLPLVTCSTD---HKTA
MUL_1725|M.ulcerans_Agy99 RQSTNQYIQMVALQFEATQCGK-DDILAGNDDPKLPLVTCSTD---HKTA
MLBr_00487|M.leprae_Br4923 RQSTKQSIQFLALQFESTHCDK-DDILAGNDDPNLPLATCSTD---HNMA
MAV_3468|M.avium_104 RQSTRQGIQFLALQFQATRCDK-DDILAGNDDPAQPLITCSTD---HKVA
Mvan_2583|M.vanbaalenii_PYR-1 RQSTEQAIQILALQFQATRCGQ-EDVLAGNDDPNLPLVTCSQD---GNEV
Mflv_3816|M.gilvum_PYR-GCK RQSSDQMIQILAMQLQASQCGQ-EDVLAGNDDPNLPLITCSQD---GTTV
TH_1363|M.thermoresistible__bu -------------------------VLAGNDDPELPLITCSTD---GQTV
MSMEG_2961|M.smegmatis_MC2_155 RQSTDPTMQILGLQVQANLCGE-DDILAGNDDPNLPLITCSTD---GSTV
MAB_2880c|M.abscessus_ATCC_199 RQSEDPRIQMLALQVQASRCYDPDDPLAGNDDPNLPLVTCGDDSQGGKAA
******* ** **. * .
Mb2618c|M.bovis_AF2122/97 YLLAPSIISGDQIQNATSGMD-QRGIGYVVDLQFKGPAANIWADYTAAH-
Rv2587c|M.tuberculosis_H37Rv YLLAPSIISGDQIQNATSGMD-QRGIGYVVDLQFKGPAANIWADYTAAH-
MMAR_2120|M.marinum_M YLLAPSIISGDQIQNATSGMD-QRSIGYVVDLQFKSQAANTWADYTAAH-
MUL_1725|M.ulcerans_Agy99 YLLAPSIISGDQIQNATSGMD-QRSIGYVVDLQFKSQAANTWADYTAAH-
MLBr_00487|M.leprae_Br4923 YLLAPSIISGDQIQNSTSGMN-QRGVGYVVDLQFKSAAADVWADFTAAH-
MAV_3468|M.avium_104 YLLGPSIISGDQIKDASASQN-QRGIGFVVDLQFKPAAANTWADFTAAH-
Mvan_2583|M.vanbaalenii_PYR-1 YLLDKSIISGEQIANASSGLN-QQAGEYVVDVEFKSDAAKVWADFTAAN-
Mflv_3816|M.gilvum_PYR-GCK YLLDKSIIAGEEIANATSGLN-QQAGEYVVDLEFKDQGSKTWAEFTAAN-
TH_1363|M.thermoresistible__bu YLLNKSIISGEQIENASSGRDPQRGGEYVVDLQFKGDATDIWADFTAAN-
MSMEG_2961|M.smegmatis_MC2_155 YLLNKSIMSGEEIKNATSGLD-QQQGQYVVDVEFKSGGAQTWADFTAAN-
MAB_2880c|M.abscessus_ATCC_199 YLLDKSIISGEEIKDASSGLD-QQRGIYVVSMEFKPTGAKVWADYTSAHW
*** **::*::* ::::. : *: :**.::** .:. **::*:*:
Mb2618c|M.bovis_AF2122/97 -IGTQTAFTLDSQVVSAPQIQEAIPGGRTQISGGDPPFTAATARQLANVL
Rv2587c|M.tuberculosis_H37Rv -IGTQTAFTLDSQVVSAPQIQEAIPGGRTQISGGDPPFTAATARQLANVL
MMAR_2120|M.marinum_M -IGTQTAFTLDSQVVSAPQILEAIPGGRTQISGGEPPFSASTARQLANVL
MUL_1725|M.ulcerans_Agy99 -IGTQTAFTLDSQVVSAPQILEAIPGGRTQISGGEPPFSASTARQLANVL
MLBr_00487|M.leprae_Br4923 -IGTQTAFTLDSEVVSVPVINEAILGGRTQISGGDPPFTAATARQLANVL
MAV_3468|M.avium_104 -IGTQTAFTLDSQVVSAPMIQEAIPGGRTQISGGDPPFTAATAKQLANVL
Mvan_2583|M.vanbaalenii_PYR-1 -VGTQTAFVLDSQVVSAPVIQEAIPGGRTQITG---QFTQTTARELANVL
Mflv_3816|M.gilvum_PYR-GCK -VGVQTAFVLDSKVVSAPVINEPIPGGKTQITG---QFTQDTARELANVL
TH_1363|M.thermoresistible__bu -VGTQTAFVLDSKVVSAPQIQEPIPGGRTQITG---RFNAETAKELANVL
MSMEG_2961|M.smegmatis_MC2_155 -VGTQTAFTLDSKVVSAPEIREAIPGGNTQITG---QFTADSAKELANVL
MAB_2880c|M.abscessus_ATCC_199 QQGTQTAFTLDSKVISAPAIREPIPGGKTEISG---QFTSDSAKQLAAAL
*.****.***:*:*.* * *.* **.*:*:* *. :*::** .*
Mb2618c|M.bovis_AF2122/97 KYGSLPLSFEPSEAQTVSATLGLSSLRAGMIAGAIGLLLVLVYSLLYYRV
Rv2587c|M.tuberculosis_H37Rv KYGSLPLSFEPSEAQTVSATLGLSSLRAGMIAGAIGLLLVLVYSLLYYRV
MMAR_2120|M.marinum_M KYGSLPLSFESSEAQTVSATLGLTSLRAGLIAGAIGLALVLLYSLLYYRV
MUL_1725|M.ulcerans_Agy99 KYGSLPLSFESSEAQTVSATLGLTSLRAGLIAGAIGLALVLLYSLLYYRV
MLBr_00487|M.leprae_Br4923 KYGSLPLSFEPSEAQTVSATLGLTSLRAGLIAGAIGLSLVLLYSLLYYRV
MAV_3468|M.avium_104 KYGSLPLSFQSSEAQTVSATLGLTSLRAGLIAGAIGLLLVLLYSLLYYRV
Mvan_2583|M.vanbaalenii_PYR-1 KYGSLPLSFESSEAETVSATLGLSSLRAGLIAGVVGLVAVLLYSLLYYRV
Mflv_3816|M.gilvum_PYR-GCK KYGSLPLSFESSEAETVSATLGLSSLRAGLIAGAVGLAAVLLYSLLYYRA
TH_1363|M.thermoresistible__bu KYGSLPLSFEASEAETVSPTLGLNSLRAGLIAGAVGLAAVLLYSLLYYRL
MSMEG_2961|M.smegmatis_MC2_155 KYGSLPLSFESSEAETVSATLGLSSLRAGLIAGAIGLAAVLLYSLLYYRV
MAB_2880c|M.abscessus_ATCC_199 KYGSLPLSFESSDAQTVSATLGLTSLKAGLIAGAIGLAIVLLYSLIYYRV
*********:.*:*:***.****.**:**:***.:** **:***:***
Mb2618c|M.bovis_AF2122/97 LGLLTALSLVASGSMVFAILVLLGRYINYTLDLAGIAGLIIGIGTTADSF
Rv2587c|M.tuberculosis_H37Rv LGLLTALSLVASGSMVFAILVLLGRYINYTLDLAGIAGLIIGIGTTADSF
MMAR_2120|M.marinum_M LGLLTALSLIASGAMVFAILVLLGRYINYTLDLAGIAGLIIGIGTTADSF
MUL_1725|M.ulcerans_Agy99 LGLLTALSLIASGAMVFAILVLLGRYINYTLDLAGIAGLIIGIGTTADSF
MLBr_00487|M.leprae_Br4923 LGLLTAFSLFCSGTIIFAILVLLGRYINYTLDLAGIAGLIIGIGTTADSF
MAV_3468|M.avium_104 LGLLTALSLAASGAMVFAILILLGRYINYTLDLAGIAGLIIGIGTTADSF
Mvan_2583|M.vanbaalenii_PYR-1 LGVLIALSLTASGAMVFAILVLLGRYISYTLDLAGIAGLIIGIGMTADSF
Mflv_3816|M.gilvum_PYR-GCK LGVLIALSLVASGAMVFAILVLLGRYINYTLDLAGIAGLIIGIGMTADSF
TH_1363|M.thermoresistible__bu LGVLTALSLVAAGALVYAVLVLLGRYINYTLDLAGVAGLIIGIGTTADSF
MSMEG_2961|M.smegmatis_MC2_155 LGLLVALSLVAAGAVVYAILVLLGRYIGYTLDLAGIAGLIIGIGTTADSF
MAB_2880c|M.abscessus_ATCC_199 LGLLTALSLVASGAMVFAILVLLGRYISYTLDLAGIAGLIIGIGTTADSF
**:* *:** .:*::::*:*:******.*******:******** *****
Mb2618c|M.bovis_AF2122/97 VVFFERIKDEIREGRSFRSAVPRGWARARKTIVSGSAVTFLAAAVLYFLA
Rv2587c|M.tuberculosis_H37Rv VVFFERIKDEIREGRSFRSAVPRGWARARKTIVSGNAVTFLAAAVLYFLA
MMAR_2120|M.marinum_M VVFFERIKDEIREGRSFRSAVPRGWARARKTIVSGNAVTFLAAAVLYFLA
MUL_1725|M.ulcerans_Agy99 VVFFERIKDEIREGRSFRSAVPRGWARARKTIVSGNAVTFLAAAVLYFLA
MLBr_00487|M.leprae_Br4923 VVFFERIKDEIREGRSFRSAVPRGWVRARKTIVSGNAVTFLAAAVLHFLA
MAV_3468|M.avium_104 VVFFERIKDEIREGRTFRSAVPRGWTRARKTIVSGNAVTFLAAAVLYFLA
Mvan_2583|M.vanbaalenii_PYR-1 VVFFERIKDEIREGRSFRSAVPRGWARARKTIVSGNAVTLLAAAVLYFLA
Mflv_3816|M.gilvum_PYR-GCK VVFFERIKDEIREGRSFRSAVPRGWARARKTILSGNAVTFLAAAVLYFLA
TH_1363|M.thermoresistible__bu VVFFERIKDEIREGRSFRSAVPRGWARARKTILSGNAVTFLAAAVLYFLA
MSMEG_2961|M.smegmatis_MC2_155 VVFFERIKDEIREGRSFRSAVPRGWVRARKTIVSGNAVTFLAAAVLYVLA
MAB_2880c|M.abscessus_ATCC_199 VVFFERIKDEIREGRSYRSAVPRGWARARKTILSGNAVTLLAAVVLYILA
***************::********.******:**.***:***.**:.**
Mb2618c|M.bovis_AF2122/97 IGQVKGFAFTLGLTTILDLVVVFLVTWPLVYLASKSSLLAKPAYNGLGAV
Rv2587c|M.tuberculosis_H37Rv IGQVKGFAFTLGLTTILDLVVVFLVTWPLVYLASKSSLLAKPAYNGLGAV
MMAR_2120|M.marinum_M IGQVKGFAFTLGLTTILDIVVVFLVTWPLVYLASKSPRLAKPAYNGLGAV
MUL_1725|M.ulcerans_Agy99 IGQVKGFAFTLGLTTILDIVVVFLVTWPLVYLASKSPRLAKPAYNGLGAV
MLBr_00487|M.leprae_Br4923 IGQVKGFAFTLGLTTILDLVVVFLVTWPLVYLASKSPLLARPAYNGLGAV
MAV_3468|M.avium_104 IGQVKGFAFTLGLTTILDLVVVFLVTWPLVYLASKSPTLAKPAYNGLGAV
Mvan_2583|M.vanbaalenii_PYR-1 VGQVKGFAFTLGLTTILDIVVVFLVTWPLVYMASKSTLWAKPSLNGLGAV
Mflv_3816|M.gilvum_PYR-GCK VGQVKGFAFTLGLTTILDVVVVFLVTWPLVYLASKSPLWAKPSLNGLGAV
TH_1363|M.thermoresistible__bu IGQVKGFAFTLGLTTIFDVMVVFLVTWPLVYLASKSPTLAKPSLNGLGAV
MSMEG_2961|M.smegmatis_MC2_155 IGQVKGFAFTLGLTTILDLVVVFLVTWPLVYLASKSPTMAKPKLNGLGAV
MAB_2880c|M.abscessus_ATCC_199 IGQVKGFAFTLGLTTILDVVVVFLVTWPLVYLSSKSATLSKPAYNGLGAV
:***************:*::***********::***. ::* ******
Mb2618c|M.bovis_AF2122/97 QQVARERRA--------MARTGRG-
Rv2587c|M.tuberculosis_H37Rv QQVARERRA--------MARTGRG-
MMAR_2120|M.marinum_M QQVARERRASDSRTRSGIAKTGRG-
MUL_1725|M.ulcerans_Agy99 QQVARERRASDSRTRSGIAKTGRE-
MLBr_00487|M.leprae_Br4923 QQVARERRAS--------AKTGRG-
MAV_3468|M.avium_104 QQVARERRAA------AHVSTGQG-
Mvan_2583|M.vanbaalenii_PYR-1 QQIARERR-------AAASAAGRG-
Mflv_3816|M.gilvum_PYR-GCK QQIARERR-------AAATAAGRG-
TH_1363|M.thermoresistible__bu QQIARERR-------AARATTGQGI
MSMEG_2961|M.smegmatis_MC2_155 QQIARERRE------VSHATAGRG-
MAB_2880c|M.abscessus_ATCC_199 QQIARERKA------AAHA------
**:****: