For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSTIGGMVALTDVLTDVPTTTAGALELFDSCPAIETDFMLGTWHGVELPTGHRLDGLLAASGWWGKHFKD CETVHPLLFPTADGTGLWPLNPVLAFAGLGAAARLPALGSRGFAGVITALKPALRARGPKARLRTTRYRG VDTATMIYDQLPVNDVFRSIDDRTVLGAMDLRGIRTPYFFVLQRDNSLRLT*
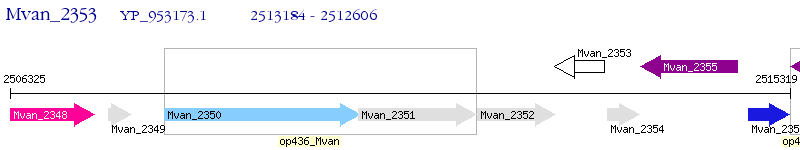
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2353 | - | - | 100% (192) | hypothetical protein Mvan_2353 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4020 | - | 9e-72 | 68.39% (193) | hypothetical protein Mflv_4020 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0274c | - | 3e-35 | 45.40% (174) | hypothetical protein MAB_0274c |
| M. marinum M | MMAR_5327 | - | 2e-13 | 32.45% (151) | hypothetical protein MMAR_5327 |
| M. avium 104 | MAV_0249 | - | 4e-16 | 34.90% (149) | hypothetical protein MAV_0249 |
| M. smegmatis MC2 155 | MSMEG_6352 | - | 9e-13 | 30.66% (137) | hypothetical protein MSMEG_6352 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4408 | - | 1e-12 | 32.61% (138) | hypothetical protein MUL_4408 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5327|M.marinum_M ---MS-ARDTFTELKNRMGAIPDAELDDFWATLEPATIDGMIGE-WKGGE
MUL_4408|M.ulcerans_Agy99 ---MN-ARDTFTEFKNRVGAIPDAELDDFWATLEPASIDGMIGE-WKGGE
MAV_0249|M.avium_104 ---MDLARKKFTEFKERSGDIPDSELDDFWAELAPATIEEMLGE-WKGGE
MSMEG_6352|M.smegmatis_MC2_155 ---MTDARGTFEAFKNRDGEIPDAELDAFWALLRPAGIDFMLGE-WKGGE
Mvan_2353|M.vanbaalenii_PYR-1 MSSTIGGMVALTDVLT-DVPTTTAGALELFDSCPAIETDFMLGT-WHGVE
Mflv_4020|M.gilvum_PYR-GCK MSSRLADMAALSEVLPDDVPTTTADALALFDSLAAVDPQFMIGT-WHGAE
MAB_0274c|M.abscessus_ATCC_199 -------MTSTSDLFALEHGTTTQEALTFFDNLEPIRAEELIGQRWHGGE
. . :: . : ::* *:* *
MMAR_5327|M.marinum_M FDTGHKMNGQLEKARWFGKTFASPTDVQPLVCLDADGN------------
MUL_4408|M.ulcerans_Agy99 FDTGHKMNGQLEKARWFGKTFTSPTDVQPLVCLDADGQ------------
MAV_0249|M.avium_104 FRTGHKMNGQLEKAGWFGKTFKSARDVQPLVCLDADGN------------
MSMEG_6352|M.smegmatis_MC2_155 FYTGHKANGFMKKLNWFGKTFVSASEAKPLVCLDADGN------------
Mvan_2353|M.vanbaalenii_PYR-1 LPTGHRLDGLLAASGWWGKHFKDCETVHPLLFPTADGTGLWPLNPVLAFA
Mflv_4020|M.gilvum_PYR-GCK LPTGHPLDGILAASGWWGKRFRDNETVDPLLFPTADGAGLFALNPAPAFA
MAB_0274c|M.abscessus_ATCC_199 VPTGHSMDGILTVSGWYGKQFDDVDHVHPLLFSDGGGD-IFAVDPMLAPM
. *** :* : *:** * . ..**: ..*
MMAR_5327|M.marinum_M --------------------------KFSNVAMGKGEASLWLEDFRGETT
MUL_4408|M.ulcerans_Agy99 --------------------------KFSNVAMGKGEASLWLEDFRGETT
MAV_0249|M.avium_104 --------------------------KFSNVELGKGEASLWLEEFRGEVT
MSMEG_6352|M.smegmatis_MC2_155 --------------------------KFSNTEAMGGEASLWLEEFRGEVT
Mvan_2353|M.vanbaalenii_PYR-1 GLG----AAARLPALGSRGFAGVITALKPALRARGPKARLRTTRYRGVDT
Mflv_4020|M.gilvum_PYR-GCK GLG----LFTRFPMLKKQNFTAAVATLRPALQARGPKARLRTTRYRGVDT
MAB_0274c|M.abscessus_ATCC_199 GLAGRISGLTESPMVKRTSPT-ALSALKPLIRTTKHRARLRNIEFRGAVS
. .* * :** :
MMAR_5327|M.marinum_M ATMVYDGQPVHDHFKRIDDD----AVMGIMNGKGVLDSG----RYYYFYL
MUL_4408|M.ulcerans_Agy99 ATMVYDGQPVHDHFKRIDDD----AVMGIMNGKGVLDNG----RYY----
MAV_0249|M.avium_104 ATMVYDGQPVHDHFKKIDDN----AVMGIMNGKGVRDNG----KYYYFYL
MSMEG_6352|M.smegmatis_MC2_155 ASMVYDGRPVHDHFKVVDDN----AVIGIMNGKGALDLSSGTPRHLYFYL
Mvan_2353|M.vanbaalenii_PYR-1 ATMIYDQLPVNDVFRSIDDR----TVLGAMDLRGIRTPY-------FFVL
Mflv_4020|M.gilvum_PYR-GCK ATMIYDQLPINDVFRLVDDTPGSEVVLGAMDLRGYDAPY-------FFVL
MAB_0274c|M.abscessus_ATCC_199 AAMVYDHKAIIDHFRKVDDA----TLLGIMDLRDMDQPY-------FFTL
*:*:** .: * *: :** .::* *: :.
MMAR_5327|M.marinum_M ERV------
MUL_4408|M.ulcerans_Agy99 ---------
MAV_0249|M.avium_104 ERV------
MSMEG_6352|M.smegmatis_MC2_155 ERI------
Mvan_2353|M.vanbaalenii_PYR-1 QRDNSLRLT
Mflv_4020|M.gilvum_PYR-GCK LRDGSRS--
MAB_0274c|M.abscessus_ATCC_199 RRD------