For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTDARGTFEAFKNRDGEIPDAELDAFWALLRPAGIDFMLGEWKGGEFYTGHKANGFMKKLNWFGKTFVSA SEAKPLVCLDADGNKFSNTEAMGGEASLWLEEFRGEVTASMVYDGRPVHDHFKVVDDNAVIGIMNGKGAL DLSSGTPRHLYFYLERI
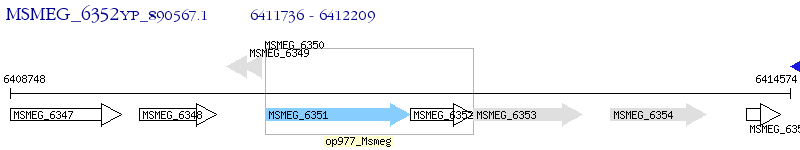
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6352 | - | - | 100% (157) | hypothetical protein MSMEG_6352 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4020 | - | 2e-11 | 29.01% (162) | hypothetical protein Mflv_4020 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00121 | - | 1e-08 | 54.00% (50) | hypothetical protein MLBr_00121 |
| M. abscessus ATCC 19977 | MAB_4387 | - | 2e-37 | 51.75% (143) | hypothetical protein MAB_4387 |
| M. marinum M | MMAR_5327 | - | 2e-59 | 68.83% (154) | hypothetical protein MMAR_5327 |
| M. avium 104 | MAV_0249 | - | 7e-60 | 69.48% (154) | hypothetical protein MAV_0249 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4408 | - | 6e-57 | 71.22% (139) | hypothetical protein MUL_4408 |
| M. vanbaalenii PYR-1 | Mvan_2353 | - | 6e-13 | 30.66% (137) | hypothetical protein Mvan_2353 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5327|M.marinum_M ---MS-ARDTFTELKNRMGAIPDAELDDFWATLEPATIDGMIGEWKGGEF
MUL_4408|M.ulcerans_Agy99 ---MN-ARDTFTEFKNRVGAIPDAELDDFWATLEPASIDGMIGEWKGGEF
MAV_0249|M.avium_104 ---MDLARKKFTEFKERSGDIPDSELDDFWAELAPATIEEMLGEWKGGEF
MSMEG_6352|M.smegmatis_MC2_155 ---MTDARGTFEAFKNRDGEIPDAELDAFWALLRPAGIDFMLGEWKGGEF
MLBr_00121|M.leprae_Br4923 ---MVWHMTHSPSSKN-TRACPAT-----------------LGEQKSDEF
MAB_4387|M.abscessus_ATCC_1997 -MDAHKARTRFTELRR-QRDINPTELDEIWAALDTVRIEDILGSWKGDEF
Mflv_4020|M.gilvum_PYR-GCK MSSRLADMAALSEVLPDDVPTTTADALALFDSLAAVDPQFMIGTWHGAEL
Mvan_2353|M.vanbaalenii_PYR-1 MSSTIGGMVALTDVLT-DVPTTTAGALELFDSCPAIETDFMLGTWHGVEL
: :* :. *:
MMAR_5327|M.marinum_M DTGHKMNGQLEKARWFGKTFASPTDVQPLVCLDADGNKFSNV--------
MUL_4408|M.ulcerans_Agy99 DTGHKMNGQLEKARWFGKTFTSPTDVQPLVCLDADGQKFSNV--------
MAV_0249|M.avium_104 RTGHKMNGQLEKAGWFGKTFKSARDVQPLVCLDADGNKFSNV--------
MSMEG_6352|M.smegmatis_MC2_155 YTGHKANGFMKKLNWFGKTFVSASEAKPLVCLDADGNKFSNT--------
MLBr_00121|M.leprae_Br4923 HARHKINDQSEKASWFGKTFKSAACILSLVCVDA-GNKDNNA--------
MAB_4387|M.abscessus_ATCC_1997 HTGHRMNGQLAAARWYGKVFKSINDVEPIVCYDNDGKLFSNN--------
Mflv_4020|M.gilvum_PYR-GCK PTGHPLDGILAASGWWGKRFRDNETVDPLLFPTADGAGLFALNPAPAFAG
Mvan_2353|M.vanbaalenii_PYR-1 PTGHRLDGLLAASGWWGKHFKDCETVHPLLFPTADGTGLWPLNPVLAFAG
: * :. *:** * . .:: *
MMAR_5327|M.marinum_M --------------------------AMGKGEASLWLEDFRGETTATMVY
MUL_4408|M.ulcerans_Agy99 --------------------------AMGKGEASLWLEDFRGETTATMVY
MAV_0249|M.avium_104 --------------------------ELGKGEASLWLEEFRGEVTATMVY
MSMEG_6352|M.smegmatis_MC2_155 --------------------------EAMGGEASLWLEEFRGEVTASMVY
MLBr_00121|M.leprae_Br4923 --------------------------GTL---------------------
MAB_4387|M.abscessus_ATCC_1997 --------------------------ELSLGGATLWEVEFRGEVTATMVY
Mflv_4020|M.gilvum_PYR-GCK LGLFTRFPMLKKQNFTAAVATLRPALQARGPKARLRTTRYRGVDTATMIY
Mvan_2353|M.vanbaalenii_PYR-1 LGAAARLPALGSRGFAGVITALKPALRARGPKARLRTTRYRGVDTATMIY
MMAR_5327|M.marinum_M DGQPVHDHFKRIDDDAVMGIMNGKGVLDSG----RYYYFYLERV------
MUL_4408|M.ulcerans_Agy99 DGQPVHDHFKRIDDDAVMGIMNGKGVLDNG----RYY-------------
MAV_0249|M.avium_104 DGQPVHDHFKKIDDNAVMGIMNGKGVRDNG----KYYYFYLERV------
MSMEG_6352|M.smegmatis_MC2_155 DGRPVHDHFKVVDDNAVIGIMNGKGALDLSSGTPRHLYFYLERI------
MLBr_00121|M.leprae_Br4923 --------------EALIGSMNGKDILDSS--------------------
MAB_4387|M.abscessus_ATCC_1997 DGQPVFDHFKRVDGSTLFGIMNGKRQRQSG----SYLYFLLERDQ-----
Mflv_4020|M.gilvum_PYR-GCK DQLPINDVFRLVDDTPGSEVVLGAMDLRGYD---APYFFVLLRDGSRS--
Mvan_2353|M.vanbaalenii_PYR-1 DQLPVNDVFRSIDDR----TVLGAMDLRGIR---TPYFFVLQRDNSLRLT
: *