For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTVDHVAMHMSDGIVNAPISVLFGIIAVVALGICAWRAREELDERTVPLAGLVAAFIFAVQMVNFPILPG VSGHLLGGALAAILVGPFTGALCVAIVLIVQAFLFADGGVTALGANITNMSVFGVAAGYASALLLYRLAR RGRDDVAVPVVGGIAFVSAVIGTVCAAMGFVLEYSLGGAATTSLGAVAGYMFGTHVLIGVGEGIITALTV MAVVRARPDLVYLLRTNRVRTAVPA
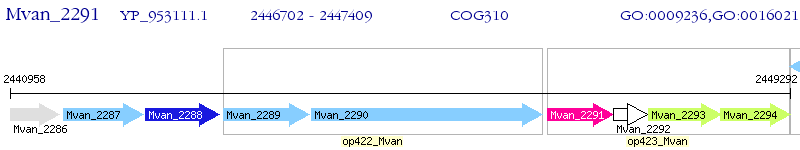
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2291 | - | - | 100% (235) | cobalamin (vitamin B12) biosynthesis CbiM protein |
| M. vanbaalenii PYR-1 | Mvan_3228 | - | 4e-05 | 30.11% (186) | BioY protein |
| M. vanbaalenii PYR-1 | Mvan_1872 | - | 5e-05 | 24.58% (240) | NADH dehydrogenase subunit N |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4058 | - | 1e-108 | 83.76% (234) | cobalamin (vitamin B12) biosynthesis CbiM protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2607 | cbiM | 5e-91 | 74.12% (228) | cobalamin biosynthesis protein CbiM |
| M. thermoresistible (build 8) | TH_3322 | - | 2e-16 | 32.40% (179) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2291|M.vanbaalenii_PYR-1 MTVDHVAMHMSDGIVNAPISVLFGIIAVVALGICAWRAREELDER--TVP
Mflv_4058|M.gilvum_PYR-GCK MSAHYVAMHMSDGIINAPISMLFVAIAVAALGICAWRAREELDER--TVP
MSMEG_2607|M.smegmatis_MC2_155 -------MHMSDGIVNVQTSLVFGALAVAALGVCALRARDELDER--TVP
TH_3322|M.thermoresistible__bu -------MHIAEGFLPPVHAVAWTVAAAPFVIYGARQVVITVRRQPSSGP
**:::*:: :: : *. : * :. : .: : *
Mvan_2291|M.vanbaalenii_PYR-1 LAGLVAAFIFAVQMVNFPILPGVSGHLLGGALAAILVGPFTGALCVAIVL
Mflv_4058|M.gilvum_PYR-GCK LAGLVAAFVFAVQMVNFPILPGVSGHLLGGALAAILVGPYTGALCVSIVL
MSMEG_2607|M.smegmatis_MC2_155 LAGLVAAFIFAVQMVNFPILPGVSGHLLGGALAAILVGPYTGALCIAIVL
TH_3322|M.thermoresistible__bu LLAAVGAFSFVMSAIKLPSVTGSSSHPTGTGLGTAIFRPPVMAFLGSIVL
* . *.** *.:. :::* :.* *.* * .*.: :. * . *: :***
Mvan_2291|M.vanbaalenii_PYR-1 IVQAFLFADGGVTALGANITNMSVFGVAAGYASALLLYRLARRGRDDVAV
Mflv_4058|M.gilvum_PYR-GCK IVQALLFADGGVTALGTNITNMSVIGVAAGYGTAVLLYRVLRRRRADLAV
MSMEG_2607|M.smegmatis_MC2_155 VVQSLLFADGGVSALGTNITNMALIGVAAGYATAVVLSTVLRGRGAAGSP
TH_3322|M.thermoresistible__bu LFQALLLAHGGITTLGANAFSMAIAGPWAGFAVYVALRRAGVG-----VL
:.*::*:*.**:::**:* .*:: * **:. : *
Mvan_2291|M.vanbaalenii_PYR-1 P--VVGGIAFVSAVIGTVCAAMGFVLEY-SLGGAATTSLGAVAGYMFGTH
Mflv_4058|M.gilvum_PYR-GCK P--VVGVIAFVSAVVGTVCAAMGFVAQY-ALGGAATTSLGTVAAYMFGTH
MSMEG_2607|M.smegmatis_MC2_155 PDRHIGVVAFVAALVGTVCAAMGFVVEY-VIGGVATTSIQAVAGYMFGTH
TH_3322|M.thermoresistible__bu P--AVFALAFVADLVTYIVTSIQLALAFPAAEGGFTESLVEFLGVFALTQ
* : :***: :: : ::: :. : * * *: . . : *:
Mvan_2291|M.vanbaalenii_PYR-1 VLIGVGEGIITALTVMAVVRARPDLVYLLRTNRVRTAVPA----------
Mflv_4058|M.gilvum_PYR-GCK VLIGIGEGVITALTVMAVVRARPDLVYLLRSHPARQAVPV----------
MSMEG_2607|M.smegmatis_MC2_155 VLIGIGEGIITALTVTAVARVRPDLVYLLRRQ--RQEVAA----------
TH_3322|M.thermoresistible__bu VPLAVVEGLITVVLVRILMHVASSELLRLRFLKSHEVPAGADVEPGSERE
* :.: **:**.: * : :. .. : ** : .
Mvan_2291|M.vanbaalenii_PYR-1 ----
Mflv_4058|M.gilvum_PYR-GCK ----
MSMEG_2607|M.smegmatis_MC2_155 ----
TH_3322|M.thermoresistible__bu EVQR