For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSEPAVDNVDRLQRSSRDMSNVPAALSQWLSTLIPDITPEITVESGIDANGMSSETILLRGRWLQAGDPV EQRWVARVAPAEADIPVFEHYRLDHQHEVMRLVAELTDVAVPPVRWLERRGDVLGRPFFLMDRVEGIVPP DVMPYTFGDNWLFDATAEQQRQLQDRTVEVLAKLHSIPNAQQVFGFLQDVDPPGPTALHRHFGWLKNWYE FSVPDIGRSPLVERALAWLEDRFPEDVAAAEPVLVWGDSRIGNVIYQDFSPVAVLDWEMATVGPREFDVA WISFAHMVFQELTKLAGLPGMPDFLREDDVRETYRELTGVEIGDLHWFHVYSAVVWCCVFMRTGARRVHF GEIERPADIETLFYHASLLKRLIGETD*
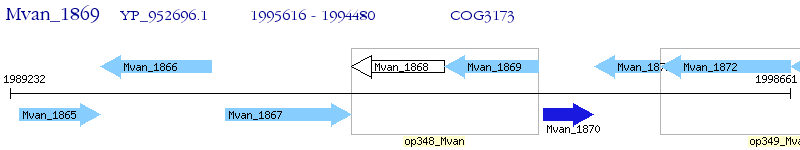
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1869 | - | - | 100% (378) | aminoglycoside phosphotransferase |
| M. vanbaalenii PYR-1 | Mvan_4201 | - | 4e-12 | 29.66% (236) | aminoglycoside phosphotransferase |
| M. vanbaalenii PYR-1 | Mvan_5603 | - | 2e-09 | 31.11% (225) | aminoglycoside phosphotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3193 | - | 1e-153 | 66.31% (377) | hypothetical protein Mb3193 |
| M. gilvum PYR-GCK | Mflv_4497 | - | 0.0 | 84.29% (382) | aminoglycoside phosphotransferase |
| M. tuberculosis H37Rv | Rv3168 | - | 1e-154 | 66.58% (377) | hypothetical protein Rv3168 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4482 | - | 3e-74 | 40.27% (375) | putative phosphotransferase |
| M. marinum M | MMAR_1393 | - | 1e-151 | 64.46% (377) | hypothetical protein MMAR_1393 |
| M. avium 104 | MAV_4058 | - | 1e-156 | 68.97% (377) | phosphotransferase enzyme family protein |
| M. smegmatis MC2 155 | MSMEG_2042 | - | 1e-164 | 71.88% (377) | phosphotransferase enzyme family protein |
| M. thermoresistible (build 8) | TH_0687 | - | 1e-148 | 73.53% (340) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2483 | - | 1e-149 | 63.93% (377) | hypothetical protein MUL_2483 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1869|M.vanbaalenii_PYR-1 MTSEPA-VDNVDRLQRSSRDMSNVPAALSQWLSTLIPDITP----EITVE
Mflv_4497|M.gilvum_PYR-GCK MTSEPA-VDNVDRLQRSSRDLSDVPAALSRWLSTKLPGVTPDAPVDITVE
TH_0687|M.thermoresistible__bu ----------------------------------------------VQVH
Mb3193|M.bovis_AF2122/97 MANEPA-IGAIDRLQRSSRDVTTLPAVISRWLSSVLPGGAAP---EVTVE
Rv3168|M.tuberculosis_H37Rv MANEPA-IGAIDRLQRSSRDVTTLPAVISRWLSSVLPGGAAP---EVTVE
MAV_4058|M.avium_104 MATEPA-VDNVDRLQRSSRDVTTVPAVMSRWLSTVLPEGAEP---EVTVE
MMAR_1393|M.marinum_M MANETP-VEDVAHLQRSSRDMTTLPAVMARWLAGVLPGNAAP---EVTVE
MUL_2483|M.ulcerans_Agy99 MANETP-VEDVAHLQRSSRDMTTLPAVMARWLAGVLPGNAAP---EVTVE
MSMEG_2042|M.smegmatis_MC2_155 MANEPA-VEDVSHLQLSSRDVSTVPVALADWLSTVLPAGAEP---RVTVE
MAB_4482|M.abscessus_ATCC_1997 MTEQTATHEDVARPAESQRDPETLRGALTDWISGQLPADAAL---SVDHA
:
Mvan_1869|M.vanbaalenii_PYR-1 SGIDANGMSSETILLRGRWL--QAG---DPVEQRWVARVAPAEADIPVFE
Mflv_4497|M.gilvum_PYR-GCK DGIDANGMSSETILLRGRWS--DGG---DPVEHRWVARVAPTDADIPVFE
TH_0687|M.thermoresistible__bu SGIDANGMSSETIVLSVDWRRRDGGRAEQRAPERYVARVAPAATDVPVFD
Mb3193|M.bovis_AF2122/97 SGVDSTGMSSETIILTARRQ--QDG---RSIQQKLVARVAPAAEDVPVFP
Rv3168|M.tuberculosis_H37Rv SGVDSTGMSSETIILTARWQ--QDG---RSIQQKLVARVAPAAEDVPVFP
MAV_4058|M.avium_104 SGVDSTGMSSETIILTARWR--QDE---RPTEQKLVARVAPTAEDVQVFP
MMAR_1393|M.marinum_M SGVDSTGMSSETIILTARWH--EDH---RPVQQKLVARVAPAAGDVPVFP
MUL_2483|M.ulcerans_Agy99 SGVDSTGMSSETIILTARWH--EDH---RPVQQKLVARVAPAAGDVPVFP
MSMEG_2042|M.smegmatis_MC2_155 DGVDSNGMSSETLLLTAEWT--EDG---APVQQRLVARVAPAPGDVPVFS
MAB_4482|M.abscessus_ATCC_1997 ELPSANGMSSETILADAQWT--ENG---QRTAHRLVIRTAPQPDSSPVFP
. .:.******:: : .: * *.** . **
Mvan_1869|M.vanbaalenii_PYR-1 HYRLDHQHEVMRLVAELTDVAVPPVRWLERRGDVLGRPFFLMDRVEGIVP
Mflv_4497|M.gilvum_PYR-GCK HYRLDHQHEVMRLVAESTDVAVPAVRWLEDTGDVLGRPFFLMDRVDGVVP
TH_0687|M.thermoresistible__bu SYRLDHQFEVMRLVSELTDVPVPRVRWLEPTGSVLGTPFFLMDHVEGVVP
Mb3193|M.bovis_AF2122/97 TYRLDHQFEVIRLVGELTDVPVPRVRWIETTGDVLGTPFFLMDYVEGVVP
Rv3168|M.tuberculosis_H37Rv TYRLDHQFEVIRLVGELTDVPVPRVRWIETTGDVLGTPFFLMDYVEGVVP
MAV_4058|M.avium_104 TYRLDHQFEVIRKVGELTDVPVPRVRWLENTGEVLGTPFFVMDYVDGLVP
MMAR_1393|M.marinum_M SYRLDHQFEVIKLVGELSDVPVPRVRWMEPTGEVLGTPFFLMDYVEGVVP
MUL_2483|M.ulcerans_Agy99 SYRLDHRFEVIKLVGELSDVPVPRVRWMVPTGEVLGTPFFLMDYVEGVVP
MSMEG_2042|M.smegmatis_MC2_155 SYRLDHQFELMRQIGELTDVPVPRVRWLESTGSVLGTPFFLMDRVDGIVP
MAB_4482|M.abscessus_ATCC_1997 SYDMRRQFDVMDTLGRHSSAVVPPVRWYCDDTAVLGGEFFVMDRVDGDVP
* : ::.::: :.. :.. ** *** *** **:** *:* **
Mvan_1869|M.vanbaalenii_PYR-1 PDVMPYTFGDNWLFDATAEQQRQLQDRTVEVLAKLHSIPNAQQVFGFLQD
Mflv_4497|M.gilvum_PYR-GCK PDVMPYTFGDNWLYDASPEQQRALQDRTVEVLAALHSIPDAQRTFGFLQE
TH_0687|M.thermoresistible__bu PDVMPYTFGDNWFYDAPAERQRELQDRTVEVLAKLHSIPQPEQTFGFLS-
Mb3193|M.bovis_AF2122/97 PDVMPYTFGDNWFADAPAERQRQLQDATVAALATLHSIPNAQNTFSFLTQ
Rv3168|M.tuberculosis_H37Rv PDVMPYTFGDNWFADAPAERQRQLQDATVAALATLHSIPNAQNTFSFLTQ
MAV_4058|M.avium_104 PDVMPYTFGGNWFADAPAERQRELQDATVGVLAKLHSIPNAANTFGFLTE
MMAR_1393|M.marinum_M PDVMPYTFGNNWFADAPAERQRELQDATVGLLAMLHSIPKPQDTFGFLSD
MUL_2483|M.ulcerans_Agy99 PDVMPYTFGNNWFADAPAERQRELQDATVGLLAMLHSIPKPQDTFGFLSD
MSMEG_2042|M.smegmatis_MC2_155 PDVMPYTFGGNWFADAPADRRRLLQDGTVAVLAKLHSIPDALNRFGFLLD
MAB_4482|M.abscessus_ATCC_1997 PDRMPYTFG-SWVTEASEADRRKMQRLTVDQLARIHAAP--VERFAFLND
** ****** .*. :*. :* :* ** ** :*: * *.**
Mvan_1869|M.vanbaalenii_PYR-1 VDPPGPTALHRHFGWLKNWYEFSVPDIGRSPLVERALAWLEDRFPEDVAA
Mflv_4497|M.gilvum_PYR-GCK IDPPGETALHRHFGWLTRWYAFSSADLGSSPLVERAISWLEEHFPHDVAA
TH_0687|M.thermoresistible__bu -GDAGRTALEQNLNRLKSWYDFAVPDIGRSPLVERALAWLDDHLPTDVAA
Mb3193|M.bovis_AF2122/97 -GRTSDTTLHRHFNWVRSWYDFAVEGIGRSPLLERTFEWLQSHWPDDAAA
Rv3168|M.tuberculosis_H37Rv -GRTSDTTLHRHFNWVRSWYDFAVEGIGRSPLLERTFEWLQSHWPDDAAA
MAV_4058|M.avium_104 -GQTGDTALRRHFNWVRSWYDFAVPDIGRSPLLERTFDWLEAHWPAAADA
MMAR_1393|M.marinum_M -GPSGDTALRRHFDWVRSWYDFAVPGIGNSALIERTFQWLQEHWPSEVAA
MUL_2483|M.ulcerans_Agy99 -GPSGDTALRRHFDWVRSWYDFAVPGIGNSALIERTFQWLQEHWPSEVAA
MSMEG_2042|M.smegmatis_MC2_155 VDPPGDTPLRRHFAWLKDWYEFAVTDIGRSPLVEHALAWLEDNFPSDVAA
MAB_4482|M.abscessus_ATCC_1997 -AREGETGLDAHVRRTHEFYEWVRGDGPTIPLIDRGFEWLRENWPEESLS
. * * :. :* : . .*::: : ** . * :
Mvan_1869|M.vanbaalenii_PYR-1 AEP----VLVWGDSRIGNVIYQDFSPVAVLDWEMATVGPREFDVAWISFA
Mflv_4497|M.gilvum_PYR-GCK SEP----VLVWGDSRIGNVIYRDFRPVAVLDWEMATIGPREFDLAWISFA
TH_0687|M.thermoresistible__bu GDP----VLAWGDARIGNVLYSDFRPVAVLDWEMATLGPRELDVAWMIFA
Mb3193|M.bovis_AF2122/97 REP----VLLWGDARVGNVLYRDFQPVAVLDWEMVALGPRELDVAWMIFA
Rv3168|M.tuberculosis_H37Rv REP----VLLWGDARVGNVLYRDFQPVAVLDWEMVALGPRELDVAWMIFA
MAV_4058|M.avium_104 AEP----VLLWGDARVGNVLYRDFVPVAVLDWEMVTLGPRELDVAWMIFA
MMAR_1393|M.marinum_M REP----VLLWGDARVGNVLYRDFRPVAVLDWEMVTLGPRELDVAWMIFA
MUL_2483|M.ulcerans_Agy99 REP----VLLWGDARVGNVLYRDFRPVAVLDWEMVTLGPRELDVAWMIFA
MSMEG_2042|M.smegmatis_MC2_155 GEP----VLAWGDSRIGNVLYEDFEPVAVLDWEMATVGPRELDVSWIIFA
MAB_4482|M.abscessus_ATCC_1997 GVGRPIDVVSWGDSRPGNVIYRDFEPVALLDWEMASVGPRELDLGWIIFL
*: ***:* ***:* ** ***:*****.::****:*:.*: *
Mvan_1869|M.vanbaalenii_PYR-1 HMVFQELTKLAGLPGMPDFLREDDVRETYRELTGVEIGDLHWFHVYSAVV
Mflv_4497|M.gilvum_PYR-GCK HMVFQELTKLAGLPGMPDFLREDDVRDTYRKLTGVEVGDLHWFHVYSAVV
TH_0687|M.thermoresistible__bu HMVFQELTGLAGLPGMPDFMREDDVRARYRELTALDLGDLHWFYVYSGVI
Mb3193|M.bovis_AF2122/97 HRVFQELAGLATLPGLPEVMREDDVRATYQALTGVELGDLHWFYVYSGVM
Rv3168|M.tuberculosis_H37Rv HRVFQELAGLATLPGLPEVMREDDVRATYQALTGVELGDLHWFYVYSGVM
MAV_4058|M.avium_104 HMVFQELAGLATLPGLPGVMREDDVRATYERLTGVEIGDLHWFYVYSGVM
MMAR_1393|M.marinum_M HMVFEELAGLATLPGLPTVMREDDVRRTYQRLTGTQLGDLHWFYVYSGVM
MUL_2483|M.ulcerans_Agy99 HMVFEELAGLATLPGLPTVMREDDVRRTYQRLTGTQLGDLHWFYVYSGVM
MSMEG_2042|M.smegmatis_MC2_155 HMVFQELAGLAGLPGLPDVLREDDVRATYTRLTGAQLGDLRWFYIYSGVI
MAB_4482|M.abscessus_ATCC_1997 HRFFQDLAEVATLPGLPDFCKREDIAAEYAALTGHHAADLDFYTAYAALQ
* .*::*: :* ***:* . :.:*: * **. . .** :: *:.:
Mvan_1869|M.vanbaalenii_PYR-1 WCCVFMRTGARRVHFGEIERPADIETLFYHASLLKRLIGETD------
Mflv_4497|M.gilvum_PYR-GCK WCCVFMRTGTRRVHFGEIERPDDIESLFYHASLLKRLIGEAD------
TH_0687|M.thermoresistible__bu WCCVFMRTGARRVHFGEIERPDDVESLFYHASLLRRLIGD--------
Mb3193|M.bovis_AF2122/97 WACVFMRTGARRVHFGEIEKPDDVESLFYHAGLMKHLLGEEH------
Rv3168|M.tuberculosis_H37Rv WACVFMRTGARRVHFGEIEKPDDVESLFYHAGLMKHLLGEEH------
MAV_4058|M.avium_104 WACVFMRTGARRVHFGETEKPDDVESLFYHAGLMKRLIGEE-------
MMAR_1393|M.marinum_M WACVFMRTGARRVHFGEVEKPDDVESLFYHAGLMKRLIGDER------
MUL_2483|M.ulcerans_Agy99 WACVFMRTGARRVHFGEVEKPDDVESLFYHAGLMKRLIGDER------
MSMEG_2042|M.smegmatis_MC2_155 WCCVFMRTGARRVRFGEIEKPEDVESLFYHASLLRRLIEEA-------
MAB_4482|M.abscessus_ATCC_1997 HAVIMARIQMRAMAFGQAQMPDDPDDLILHRATLEKMLNGTYWAGVKA
. :: * * : **: : * * : *: * . :.:::