For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSVSGAEVGEDRYAWVSKARCASADPEGLFVRGAAQKDAAKICRGCPVITECAADALDHRVEYGVWGGMT ERQRRALLKAHPEVVSWAAFLAEQKRRAEAG
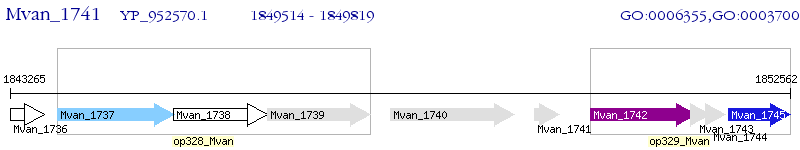
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1741 | - | - | 100% (101) | transcription factor WhiB |
| M. vanbaalenii PYR-1 | Mvan_5441 | - | 4e-36 | 72.16% (97) | transcription factor WhiB |
| M. vanbaalenii PYR-1 | Mvan_1734 | - | 1e-11 | 41.18% (68) | transcription factor WhiB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3706c | whiB4 | 3e-35 | 72.73% (88) | putative transcriptional regulatory protein whiB-like whiB4 |
| M. gilvum PYR-GCK | Mflv_1366 | - | 4e-36 | 70.71% (99) | transcription factor WhiB |
| M. tuberculosis H37Rv | Rv3681c | whiB4 | 3e-35 | 72.73% (88) | transcriptional regulatory protein WHIB-like WHIB4 |
| M. leprae Br4923 | MLBr_02307 | - | 2e-29 | 71.43% (77) | hypothetical protein MLBr_02307 |
| M. abscessus ATCC 19977 | MAB_0409 | - | 3e-35 | 67.00% (100) | putative transcriptional regulator WhiB |
| M. marinum M | MMAR_5170 | whiB4 | 2e-33 | 68.54% (89) | transcriptional regulatory protein Whib-like WhiB4 |
| M. avium 104 | MAV_0447 | - | 8e-26 | 73.53% (68) | transcription factor WhiB |
| M. smegmatis MC2 155 | MSMEG_6199 | - | 7e-35 | 72.53% (91) | transcription factor WhiB |
| M. thermoresistible (build 8) | TH_1624 | whiB4 | 5e-34 | 75.61% (82) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN WHIB-LIKE |
| M. ulcerans Agy99 | MUL_4256 | whiB4 | 8e-22 | 71.93% (57) | transcriptional regulatory protein Whib-like WhiB4 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3706c|M.bovis_AF2122/97 MSGTRPAARRTNLTAAQN-VVRSVDAEERIAWVSKALCRTTDPDELFVRG
Rv3681c|M.tuberculosis_H37Rv MSGTRPAARRTNLTAAQN-VVRSVDAEERIAWVSKALCRTTDPDELFVRG
MMAR_5170|M.marinum_M MSGIRPVDGRANLTSAQN-LLSTGEAEERINWVSKALCRATDPDELFVRG
MUL_4256|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02307|M.leprae_Br4923 ------------------------------------MCRATDPDELFVRG
MAV_0447|M.avium_104 -----------------------------------------------MRG
Mflv_1366|M.gilvum_PYR-GCK MSSIKPAARPTTVTAPTPPVVQGKEAEARIAWVSQARCRQTDPDELFVRG
MSMEG_6199|M.smegmatis_MC2_155 MSATHTVARKASTLSTTTNTAQHRDSEARIAWVSQARCRQADPDELFVRG
MAB_0409|M.abscessus_ATCC_1997 MSASRPAMSALHTAVSID------GGEARIAWVSQARCRQGDPDELFVRG
Mvan_1741|M.vanbaalenii_PYR-1 MSVSGAEV-----------------GEDRYAWVSKARCASADPEGLFVRG
TH_1624|M.thermoresistible__bu MTST---------------------FEDRIDWVLRARCRAADPDTLFVRG
Mb3706c|M.bovis_AF2122/97 AAQRKAAVICRHCPVMQECAADALDNKVEFGVWGGMTERQRRALLKQHPE
Rv3681c|M.tuberculosis_H37Rv AAQRKAAVICRHCPVMQECAADALDNKVEFGVWGGMTERQRRALLKQHPE
MMAR_5170|M.marinum_M AAQRKAAVICRHCPVMQECGADALDNKVEFGVWGGMTERQRRALLKQHPE
MUL_4256|M.ulcerans_Agy99 -------MICRHCPVMQECGADALDNKVEFGVWGGMTERQRRALLKQHPE
MLBr_02307|M.leprae_Br4923 AAQRKAAVICRHCPVMQECRADALDNKVEFGVWGGMTERQRRALLKQHPE
MAV_0447|M.avium_104 AAQRKAAVICRHCPVMQECGADALDNRVEFGVWGGMTERQRRALLKQHPE
Mflv_1366|M.gilvum_PYR-GCK AAQRKAAVICRHCPVIAECGADALDNRVEFGVWGGMTERQRRALLKQHPE
MSMEG_6199|M.smegmatis_MC2_155 AAQRKAAVICRHCPVILECGADALDNRVEFGVWGGMTERQRRALLKQHPE
MAB_0409|M.abscessus_ATCC_1997 AAQRKAAVICRHCPVMMECGADALDNRVEFGVWGGLTERQRRAMLKAHPE
Mvan_1741|M.vanbaalenii_PYR-1 AAQKDAAKICRGCPVITECAADALDHRVEYGVWGGMTERQRRALLKAHPE
TH_1624|M.thermoresistible__bu AAQRKAAKICRPCPVRTQCAADALDNRIEYGVWGGLTERQRRALLRRHPE
*** *** :* *****:::*:*****:*******:*: ***
Mb3706c|M.bovis_AF2122/97 VVSWSDYLE-KRKRRTGTAG
Rv3681c|M.tuberculosis_H37Rv VVSWSDYLE-KRKRRTGTAG
MMAR_5170|M.marinum_M VVSWSDYFE-KRKRRS--VG
MUL_4256|M.ulcerans_Agy99 VVSWSDYFE-KRKRRS--VG
MLBr_02307|M.leprae_Br4923 VVSWADFFD-TRKHRN--VS
MAV_0447|M.avium_104 VVSWADFFD-KRRNRS--AG
Mflv_1366|M.gilvum_PYR-GCK VVSWADFFAAQRKHRT--AG
MSMEG_6199|M.smegmatis_MC2_155 VSSWADFFAAQRKHRS--AV
MAB_0409|M.abscessus_ATCC_1997 VESWADFFAAQRKHKS--AV
Mvan_1741|M.vanbaalenii_PYR-1 VVSWAAFLAEQKRRAEAG--
TH_1624|M.thermoresistible__bu VDSWAEFLP---RVMD----
* **: :: :