For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLHQSTVDTLTAWRPADPAQDSLRHAVLAFLAAREDACLRDCVPGHITGSALVLDHSGRHALLTLHPRIG RWLQLGGHCEPEDETIVAAALREATEESGIDGLTITSEPAALHVHPVTCSLGVPTRHLDVQFVVRAPEGA EITRSDESLDLRWWPIDELPRDCDFGLTQLAAAAAGSAAGYSPANVHPP*
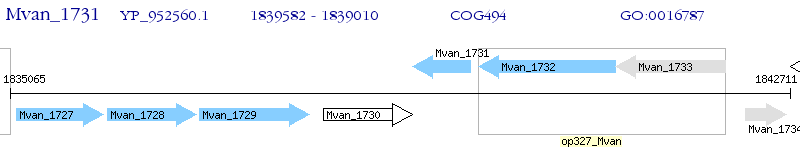
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1731 | - | - | 100% (190) | NUDIX hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4732 | - | 1e-89 | 88.07% (176) | NUDIX hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00755 | - | 1e-20 | 62.86% (70) | hypothetical protein MLBr_00755 |
| M. abscessus ATCC 19977 | MAB_3609 | - | 3e-61 | 62.43% (173) | hypothetical protein MAB_3609 |
| M. marinum M | MMAR_1278 | - | 3e-63 | 67.04% (179) | hypothetical protein MMAR_1278 |
| M. avium 104 | MAV_4227 | - | 3e-64 | 65.71% (175) | hydrolase, nudix family protein, putative |
| M. smegmatis MC2 155 | MSMEG_1827 | - | 6e-76 | 77.71% (175) | hydrolase, nudix family protein, putative |
| M. thermoresistible (build 8) | TH_0778 | - | 9e-72 | 75.15% (169) | hydrolase, nudix family protein, putative |
| M. ulcerans Agy99 | MUL_2609 | - | 4e-61 | 69.23% (169) | hypothetical protein MUL_2609 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1731|M.vanbaalenii_PYR-1 -----------MS--LHQSTVDTLTAWRPADPAQDSLRHAVLAFLAARED
Mflv_4732|M.gilvum_PYR-GCK -----------MS--LHQSTVDVLTAWNPPDPAQDSLRHSVLAFLAARED
MSMEG_1827|M.smegmatis_MC2_155 -----------MS--LHDSATRLLTHWDAPDPAQDSLRHAVLSFLAARPD
TH_0778|M.thermoresistible__bu -----------MSDPLHASAVRLLTEWRAPDPAQDSLRHAVLSFLAARPD
MMAR_1278|M.marinum_M ------MKVAPVG--VRESAIAMLSQWRAPDPAQDSLRHAVLAFLHARAD
MUL_2609|M.ulcerans_Agy99 ----------------------MLSQWRAPDPAQDSLRHAVLALLHARAD
MAV_4227|M.avium_104 -----------MS--VRDSVLALLTQWDPPDAAQDSLRHAVLAFVHARPD
MAB_3609|M.abscessus_ATCC_1997 MTARHPGEGHQMPSDLRESAIELLSSWQAPDAEQDSLRHSVLAFLDANPD
MLBr_00755|M.leprae_Br4923 --------------------------------------------------
Mvan_1731|M.vanbaalenii_PYR-1 ACLRDCVPGHITGSALVLDHSGRHALLTLHPRIGRWLQLGGHCEPEDETI
Mflv_4732|M.gilvum_PYR-GCK ACLRECAPGHITGSALVVDHRGERALLTLHPRVGRWLQLGGHCEPTDETI
MSMEG_1827|M.smegmatis_MC2_155 ACLRACVPGHITASALVIDHTGTHTLLTLHPRFGRWLQLGGHCEDTDPDI
TH_0778|M.thermoresistible__bu GCRRECAPGHITASALVVDHTGTQALLTLHPRIGRWVQLGGHCEPGDADI
MMAR_1278|M.marinum_M ACLRECEAGHITASTLVLDERG-HVLLTLHPRLGRWVQLGGHCEDDHD-I
MUL_2609|M.ulcerans_Agy99 ACLRECEAGHITASTLVLDDRG-HVLLTLHPRLGRWVQLGGHCEDDHD-I
MAV_4227|M.avium_104 ACRRECEPGHVTASTLVLDHTGDRVLLTLHRRLGRWVQLGGHCDDDDAGI
MAB_3609|M.abscessus_ATCC_1997 ACRRRSAAGHVTASALVVNHDGSQALLTLHPRVGKWLQLGGHCEEEDPTI
MLBr_00755|M.leprae_Br4923 --------------------------------------------------
Mvan_1731|M.vanbaalenii_PYR-1 VAAALREATEESGIDGLTITSEPAALHVHPVTCSLGVPTRHLDVQFVVRA
Mflv_4732|M.gilvum_PYR-GCK VAAALREATEESGISGLTIEPEPAALHVHPVTCSLGVPTRHLDVQFVVRA
MSMEG_1827|M.smegmatis_MC2_155 HAAALREATEESGIEGLTIDPDLAALHVHPVTCSLGVPTRHLDMQFLVRA
TH_0778|M.thermoresistible__bu RAAALREAAEESGITGLRVDDELAAVHVHPVTCSLGVPTRHLDLQFVVHA
MMAR_1278|M.marinum_M VAAALREATEESGIDNLLIDPELVAVHVHPVTCSLGVPTRHLDLQFVARA
MUL_2609|M.ulcerans_Agy99 VAAALREATEESGINNLLIDPELVAVHVHPVTCSLGVPTRHLDLQFVARA
MAV_4227|M.avium_104 VAAALREATEESGIDGLRMAPGLAAVHVHPVTCSLGLPTRHLDLQFVAHA
MAB_3609|M.abscessus_ATCC_1997 HAAALREATEESGIDDLLLEPNLLGIHVHPITCSLGVPTRHLDLQFLAHA
MLBr_00755|M.leprae_Br4923 --MALREATKESGVADLRITPELTTIHVHPVTCSLSEPTRHLDLQFVAHA
*****::***: .* : :****:****. ******:**:.:*
Mvan_1731|M.vanbaalenii_PYR-1 PEGAEITRSDESLDLRWWPIDELPRDCDFGLTQLAAAAAGSAAGYSPANV
Mflv_4732|M.gilvum_PYR-GCK PEGAEIVRSDESLDLRWWPLDKLPEGVDFGLTQLAAAAASLY--------
MSMEG_1827|M.smegmatis_MC2_155 PEGAEIVCSDESLDLRWWPLDALPDGTDFGLDQLAAAARRQTSVE-----
TH_0778|M.thermoresistible__bu PPAAEIACSDESLDLRWWPLDGLPAGADFGLTQLVGRFRRVHDR------
MMAR_1278|M.marinum_M PAGAQITVSEESDDLQWWPPDALPPGTDHALRYLVDRAAQRA--------
MUL_2609|M.ulcerans_Agy99 PAGAQITVSEESDDLQWWPPDALPPGTDHALRYLVDRAAQRA--------
MAV_4227|M.avium_104 PAGARIAISDESEDLRWWPVDALPAGTDHALAYLVAQATRASR-------
MAB_3609|M.abscessus_ATCC_1997 PEGAQIAVSDESLDLRWWPLDQIPE-EDHSVVVLAQRARNRLR-------
MLBr_00755|M.leprae_Br4923 LDGVHIVVSDESEALRWWPAEALRP----KLTVARKSARRSR--------
...*. *:** *:*** : : :
Mvan_1731|M.vanbaalenii_PYR-1 HPP
Mflv_4732|M.gilvum_PYR-GCK ---
MSMEG_1827|M.smegmatis_MC2_155 ---
TH_0778|M.thermoresistible__bu ---
MMAR_1278|M.marinum_M ---
MUL_2609|M.ulcerans_Agy99 ---
MAV_4227|M.avium_104 ---
MAB_3609|M.abscessus_ATCC_1997 ---
MLBr_00755|M.leprae_Br4923 ---