For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLRWAILLVVTVAVTVPLTKLGVPSAALFAALVVGMVLALMSAGPARVPRRLGVAAQGVLGVYIGTMVQQ DALSALRDDWAIVLGVAVGTLVLSVIAGALLGMRRDVTALTGSLALVAGGASGLVAIARELGGDDRVVAV VQYLRVALVTASMPVVVTVIYHADRSRAAPEAMENSTAPWYLSLAVMVALVVVGALGGRLIRLPGAGLLG PLALTVALEVSGLSFGLSVPVLLVQAGYMLIGWQAGLAFTRESLKSVGRLLPAALGLIVFIGVATAGLGI VLARVAGLTPLEGYLATSPGGVYAVLATAVETGSNVTFVIAAQVVRILLMLFAAPFLARAVVWAGRRFGK GQSRAITAESREPIRVAD
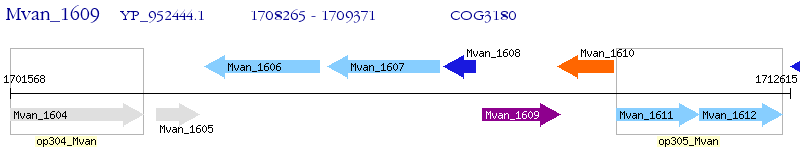
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1609 | - | - | 100% (368) | putative ammonia monooxygenase |
| M. vanbaalenii PYR-1 | Mvan_3161 | - | 4e-67 | 42.48% (339) | putative ammonia monooxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4829 | - | 1e-171 | 85.33% (368) | putative ammonia monooxygenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1698 | - | 1e-144 | 75.66% (341) | putative ammonia monooxygenase superfamily protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1609|M.vanbaalenii_PYR-1 ---------MLRWAILLVVTVAVTVPLTKLGVPSAALFAALVVGMVLALM
Mflv_4829|M.gilvum_PYR-GCK ---MKWWRSILRWTCLLAVTGAVTVPLTRVGVPSAALFAALAVGIVFALT
MSMEG_1698|M.smegmatis_MC2_155 MLKSRWSLIALRWLLLLAVTIAVTVPLTLIGVPSAALFAALVVGVVLAIA
*** **.** ******* :***********.**:*:*:
Mvan_1609|M.vanbaalenii_PYR-1 SAGPARVPRRLGVAAQGVLGVYIGTMVQQDALSALRDDWAIVLGVAVGTL
Mflv_4829|M.gilvum_PYR-GCK VGGPARVPRRLGIAAQGVLGVYIGTMVHRDALSALKGDWVIVLAVAVGTL
MSMEG_1698|M.smegmatis_MC2_155 ALAPERVPRQAGVVAQGVLGVYIGTMVHQDAVDALHSDWAIVLAIAVATL
.* ****: *:.*************::**:.**:.**.***.:**.**
Mvan_1609|M.vanbaalenii_PYR-1 VLSVIAGALLGMRRDVTALTGSLALVAGGASGLVAIARELGGDDRVVAVV
Mflv_4829|M.gilvum_PYR-GCK LLSVLAGALLGMRRDVTPLTGSLALVAGGASGLVAIARELGGDDRVVAVV
MSMEG_1698|M.smegmatis_MC2_155 VISVIAGALLGLHRDVSPLTGSLALVAGGASGLVAIARELGGDDRVVSVV
::**:******::***:.*****************************:**
Mvan_1609|M.vanbaalenii_PYR-1 QYLRVALVTASMPVVVTVIYHADRSRAAPEAMENSTAPWYLSLAVMVALV
Mflv_4829|M.gilvum_PYR-GCK QYLRVALVTASMPVVVTLIYHADRSRATTQIAETPAAPWYLSVAIMAALV
MSMEG_1698|M.smegmatis_MC2_155 QYLRVALVTATIPLVVTVVFHADRSHPAVDLPQTAAAPWYLSLAMLAGLV
**********::*:***:::*****:.: : :..:******:*::..**
Mvan_1609|M.vanbaalenii_PYR-1 VVGALGGRLIRLPGAGLLGPLALTVALEVSGLSFGLSVPVLLVQAGYMLI
Mflv_4829|M.gilvum_PYR-GCK VVGAVGGRLIRLPGAGLLGPLALTVALEVSGLSFGLSVPMLLVQAGYMLI
MSMEG_1698|M.smegmatis_MC2_155 IVGAVGGRLLHMPGAGLLGPLALTVVLQLTGLSFGLTVPDVLVQAGYMVI
:***:****:::*************.*:::******:** :*******:*
Mvan_1609|M.vanbaalenii_PYR-1 GWQAGLAFTRESLKSVGRLLPAALGLIVFIGVATAGLGIVLARVAGLTPL
Mflv_4829|M.gilvum_PYR-GCK GWQAGLAFTRASLRSVGRLLPTALALIVFIGVATAGLGVLLARVAGLTPL
MSMEG_1698|M.smegmatis_MC2_155 GWQAGVAFTRESLRAIGRILPLALLLIVVLGVATALLGVVLANVTGVTQL
*****:**** **:::**:** ** ***.:***** **::**.*:*:* *
Mvan_1609|M.vanbaalenii_PYR-1 EGYLATSPGGVYAVLATAVETGSNVTFVIAAQVVRILLMLFAAPFLARAV
Mflv_4829|M.gilvum_PYR-GCK EGYLATSPGGVYAVLATAVETGSNVTFVIAAQVVRILLMLFTAPLLARAV
MSMEG_1698|M.smegmatis_MC2_155 EGYLATSPGGVYAVLATAVETGSNVTFVIAAQVLRVLLMLFAAPLLARVM
*********************************:*:*****:**:***.:
Mvan_1609|M.vanbaalenii_PYR-1 VWAGRRFGKGQSRAITAESREPIRVAD
Mflv_4829|M.gilvum_PYR-GCK VWASRKLSPDQSPAMTADSREPIRVAD
MSMEG_1698|M.smegmatis_MC2_155 IRLTGRQTEAAEVATPARR--------
: : . * .*