For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KWWRSILRWTCLLAVTGAVTVPLTRVGVPSAALFAALAVGIVFALTVGGPARVPRRLGIAAQGVLGVYIG TMVHRDALSALKGDWVIVLAVAVGTLLLSVLAGALLGMRRDVTPLTGSLALVAGGASGLVAIARELGGDD RVVAVVQYLRVALVTASMPVVVTLIYHADRSRATTQIAETPAAPWYLSVAIMAALVVVGAVGGRLIRLPG AGLLGPLALTVALEVSGLSFGLSVPMLLVQAGYMLIGWQAGLAFTRASLRSVGRLLPTALALIVFIGVAT AGLGVLLARVAGLTPLEGYLATSPGGVYAVLATAVETGSNVTFVIAAQVVRILLMLFTAPLLARAVVWAS RKLSPDQSPAMTADSREPIRVAD*
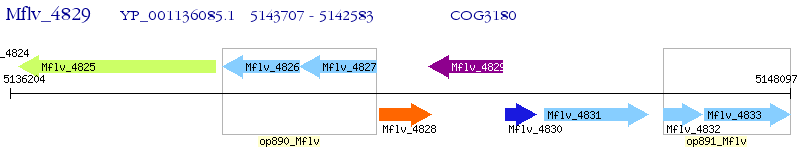
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4829 | - | - | 100% (374) | putative ammonia monooxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01562 | - | 1e-05 | 26.18% (317) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1887 | efpA | 4e-07 | 28.47% (281) | integral membrane efflux protein EfpA |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1698 | - | 1e-144 | 74.06% (347) | putative ammonia monooxygenase superfamily protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2115 | efpA | 2e-07 | 28.47% (281) | integral membrane efflux protein EfpA |
| M. vanbaalenii PYR-1 | Mvan_1609 | - | 1e-171 | 85.33% (368) | putative ammonia monooxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4829|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1609|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1698|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1887|M.marinum_M MTALNDTERAVHNWTSGRQERPTSARATRETETASERASRYYPTWLPSR-
MUL_2115|M.ulcerans_Agy99 MTALNDTERAVHNWTSGRQERPTSARATRETETASERASRYYPTWLPSR-
MLBr_01562|M.leprae_Br4923 MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSK-YPTLLPSGR
Mflv_4829|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1609|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1698|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_1887|M.marinum_M ----RFIAAVIAIGGMQLLATMDSTVAIVALPKIQDELSLSDAGRSWVIT
MUL_2115|M.ulcerans_Agy99 ----RFIAAVIAIGGMQLLATMDSTVAIVALPKIQDELSLSDAGRSWVIT
MLBr_01562|M.leprae_Br4923 FPSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDAGRSWGIT
Mflv_4829|M.gilvum_PYR-GCK ----------------------MKWWRSILRWTCLLAVTGAVTVPLTRVG
Mvan_1609|M.vanbaalenii_PYR-1 ----------------------------MLRWAILLVVTVAVTVPLTKLG
MSMEG_1698|M.smegmatis_MC2_155 -------------------MLKSRWSLIALRWLLLLAVTIAVTVPLTLIG
MMAR_1887|M.marinum_M AYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEATLV
MUL_2115|M.ulcerans_Agy99 AYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEATLV
MLBr_01562|M.leprae_Br4923 AYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDEVTLV
: : * **: . :
Mflv_4829|M.gilvum_PYR-GCK VP------SAALFAALAVGIVFALTVGGPARVPRRLGIAAQGVLGVYIGT
Mvan_1609|M.vanbaalenii_PYR-1 VP------SAALFAALVVGMVLALMSAGPARVPRRLGVAAQGVLGVYIGT
MSMEG_1698|M.smegmatis_MC2_155 VP------SAALFAALVVGVVLAIAALAPERVPRQAGVVAQGVLGVYIGT
MMAR_1887|M.marinum_M IARLSQGVGSAIASPTGLALVATTFPKGPARNAATAVFAAMTAVGSVMG-
MUL_2115|M.ulcerans_Agy99 IARLSQGVGSAIASPTGLALVATTFPKGPTRNAATAVFAAMTAVGSVMG-
MLBr_01562|M.leprae_Br4923 IARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMG-
:. .:*: :. :.:* : .. * . ..* .:* :*
Mflv_4829|M.gilvum_PYR-GCK MVHRDALSALKGDWVIVLAVAVGTLLLSVLAGALLGMRRDVTPLTGSLAL
Mvan_1609|M.vanbaalenii_PYR-1 MVQQDALSALRDDWAIVLGVAVGTLVLSVIAGALLGMRRDVTALTGSLAL
MSMEG_1698|M.smegmatis_MC2_155 MVHQDAVDALHSDWAIVLAIAVATLVISVIAGALLGLHRDVSPLTGSLAL
MMAR_1887|M.marinum_M LVVGGALTEVSWRLAFMVNVPIGLVMIYLARTALRETNRERMKLDAAGAI
MUL_2115|M.ulcerans_Agy99 LVVGGALTEVSWRLAFMVNVPIGLVMIYLARTALRETNRERMKLDAAGAI
MLBr_01562|M.leprae_Br4923 LVVGGALTEVSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLDAAGAI
:* .*: : .::: :.:. ::: : ** .:: * .: *:
Mflv_4829|M.gilvum_PYR-GCK VAGGASGLVAIARELGGD----------DRVVAVVQYLRVALVTASMP-V
Mvan_1609|M.vanbaalenii_PYR-1 VAGGASGLVAIARELGGD----------DRVVAVVQYLRVALVTASMP-V
MSMEG_1698|M.smegmatis_MC2_155 VAGGASGLVAIARELGGD----------DRVVSVVQYLRVALVTATIP-L
MMAR_1887|M.marinum_M LATLACTAAVFAFSMGPEKGWLSITTIGSGAVAFAAALGFVLVERTAENP
MUL_2115|M.ulcerans_Agy99 LATLACTAAVFAFSMGPEKGWLSITTIGSGAVAFAAALGFVLVERTAENP
MLBr_01562|M.leprae_Br4923 LATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVERTAENP
:* *. ..:* .:* : . *:.. : ..:* :
Mflv_4829|M.gilvum_PYR-GCK VVTLIYHADRSRATTQIAETPAAPWYLSVAIMAALVVVGAVGGRLIRLPG
Mvan_1609|M.vanbaalenii_PYR-1 VVTVIYHADRSRAAPEAMENSTAPWYLSLAVMVALVVVGALGGRLIRLPG
MSMEG_1698|M.smegmatis_MC2_155 VVTVVFHADRSHPAVDLPQTAAAPWYLSLAMLAGLVIVGAVGGRLLHMPG
MMAR_1887|M.marinum_M VVPFDLFHDRNRLVTFIAIFLAGGVMFSLTVCIGLYVQDVLGYSALRAGV
MUL_2115|M.ulcerans_Agy99 VVPFDLFHDRNRLVTFIAIFLAGGVMFSLTVCIGLYVQDVLGYSALRAGV
MLBr_01562|M.leprae_Br4923 VVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDILGYSALRAGI
**.. . **.: . :. :*::: .* : . :* ::
Mflv_4829|M.gilvum_PYR-GCK AGLLGPLALTVALEVSGLSFGLSVPMLLVQAG--YMLIGWQAGLAFTRAS
Mvan_1609|M.vanbaalenii_PYR-1 AGLLGPLALTVALEVSGLSFGLSVPVLLVQAG--YMLIGWQAGLAFTRES
MSMEG_1698|M.smegmatis_MC2_155 AGLLGPLALTVVLQLTGLSFGLTVPDVLVQAG--YMVIGWQAGVAFTRES
MMAR_1887|M.marinum_M GFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLVCAMIYGWAFMQRG
MUL_2115|M.ulcerans_Agy99 GFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGYLLVCAMIYGWAFMQRG
MLBr_01562|M.leprae_Br4923 GFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAMLYGWAFMNRG
. : :*: : * ::. .. * :*. .* :: . * ** . .
Mflv_4829|M.gilvum_PYR-GCK LRSVGRLLP------TALALIVFIGVATAGLGVLLARVAGLTPLEGYLAT
Mvan_1609|M.vanbaalenii_PYR-1 LKSVGRLLP------AALGLIVFIGVATAGLGIVLARVAGLTPLEGYLAT
MSMEG_1698|M.smegmatis_MC2_155 LRAIGRILP------LALLLIVVLGVATALLGVVLANVTGVTQLEGYLAT
MMAR_1887|M.marinum_M VPYFPNLVAPIVIGGIGIGMAVVPLTLAAIAGVGFDRIGPVSALTLMLQS
MUL_2115|M.ulcerans_Agy99 VPYFPNLVAPIVIGGIGIGMAVVPLTLAAIAGVGFDRIGPVSALTLMLQS
MLBr_01562|M.leprae_Br4923 VPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGPVSAIALMLQS
: . .::. .: : *. . :* *: : .: :: : * :
Mflv_4829|M.gilvum_PYR-GCK SPGGVYAVLATAVETG---------SNVTFVIAAQVVRILLMLFTAPLLA
Mvan_1609|M.vanbaalenii_PYR-1 SPGGVYAVLATAVETG---------SNVTFVIAAQVVRILLMLFAAPFLA
MSMEG_1698|M.smegmatis_MC2_155 SPGGVYAVLATAVETG---------SNVTFVIAAQVLRVLLMLFAAPLLA
MMAR_1887|M.marinum_M LGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLHALDHGYTYGLLWV
MUL_2115|M.ulcerans_Agy99 LGGPLVLAGIQAVITSRTLYLGGTTGPVKFMNDAQLHALDHGYTYGLLWV
MLBr_01562|M.leprae_Br4923 LGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRALDHGYTYGLLWV
* : . ** *. . *.*: **: : . : .
Mflv_4829|M.gilvum_PYR-GCK R--AVVWASRKLSPDQSPAMTADSREPIRVAD----
Mvan_1609|M.vanbaalenii_PYR-1 R--AVVWAGRRFGKGQSRAITAESREPIRVAD----
MSMEG_1698|M.smegmatis_MC2_155 R--VMIRLTGRQTEAAEVATPARR------------
MMAR_1887|M.marinum_M AGAAVIVGGAAVFIGYTPEQVAHAQEVKEAIDAGEL
MUL_2115|M.ulcerans_Agy99 AGAAVIVGGAAVFIGYTPEQVAHAQEVKEAIDAGEL
MLBr_01562|M.leprae_Br4923 AGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL
.:: *