For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTNASIRPGRIDATNGQVESAPDAVRAMFERHGIRTVQFGMADIDGQVRGKFVPAEFFLDSVARRGSSIP NIVFGWDIEDTLMDCFTVSGWHTGYSDVVLAPDLSTLRPVPWEPGHAFVLCDVVNTDGSPVPVAPRTVLA GQVERAAALGYRLTAGYELEFYLYRETPETAAAKGYRNLTPASPGTATFSLNRHSALEDVIGAIREGMRA CDIPVLASNTEYGAGQIEINIEHADALTTADRVAIYKNGVRRIAEQHGYLATFMAKVATDSAGSSAHIHQ SMQQLDAPARNAMWDGDRPSALMRQAVAGQLASMRDFTVLYCPTVNSYKRRVPGSWSPVSAAWGVDNRTA ALRVIAHDEATCRMENRVPGADANPYLALAACLAGVLHGIENQLEPPAAVTGNAYQGADSPLPRSLAEAV DAFGASSLARTAFGDAFVDHYLASRGWERDRHREAVSDWETGRYFSRV
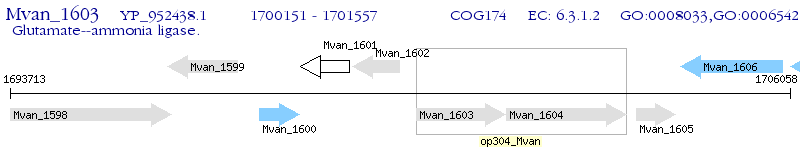
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1603 | - | - | 100% (468) | glutamate--ammonia ligase |
| M. vanbaalenii PYR-1 | Mvan_2278 | - | 2e-92 | 40.91% (462) | glutamate--ammonia ligase |
| M. vanbaalenii PYR-1 | Mvan_0612 | - | 1e-71 | 38.24% (442) | glutamate--ammonia ligase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2885c | glnA4 | 1e-91 | 43.02% (437) | glutamine synthetase |
| M. gilvum PYR-GCK | Mflv_4065 | - | 2e-94 | 42.95% (440) | glutamate--ammonia ligase |
| M. tuberculosis H37Rv | Rv2860c | glnA4 | 1e-91 | 43.02% (437) | glutamine synthetase |
| M. leprae Br4923 | MLBr_01631 | glnA2 | 5e-39 | 29.32% (457) | glutamine synthase class II |
| M. abscessus ATCC 19977 | MAB_1920 | - | 1e-40 | 30.55% (455) | glutamine synthetase |
| M. marinum M | MMAR_3294 | glnA2 | 4e-40 | 29.54% (457) | glutamine synthetase GlnA2 |
| M. avium 104 | MAV_3718 | glnA | 7e-91 | 43.28% (439) | glutamine synthetase catalytic domain |
| M. smegmatis MC2 155 | MSMEG_2595 | - | 2e-90 | 41.55% (438) | gamma-glutamylisopropylamide synthetase |
| M. thermoresistible (build 8) | TH_3396 | - | 5e-39 | 29.97% (387) | PUTATIVE glutamine synthetase, type I |
| M. ulcerans Agy99 | MUL_1335 | glnA2 | 2e-40 | 29.54% (457) | glutamine synthetase GlnA2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3294|M.marinum_M -----------------MDRQKEFVLRTLEERD---IRFVRLWFTDVLGF
MUL_1335|M.ulcerans_Agy99 -----------------MDRQKEFVLRTLEERD---IRFVRLWFTDVLGF
MLBr_01631|M.leprae_Br4923 -----------------MDRQKEFVLRTLEERD---IRFVRLWFTDVLGY
TH_3396|M.thermoresistible__bu -----------------MDRQKEFVLRSLEERD---IRFVRLWFTDVLGF
MAB_1920|M.abscessus_ATCC_1997 -----------------MDRQKEFVLRTLEERD---IRFVRLWFTDVLGY
Mb2885c|M.bovis_AF2122/97 ----------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGR
Rv2860c|M.tuberculosis_H37Rv ----------MTGPGSPPLAWTELERLVAAGD----VDTVIVAFTDMQGR
MAV_3718|M.avium_104 ---MASEDSALTGSDTAMLSLAALDRLVAAGAPETRVDTVIVAFPDMQGR
Mflv_4065|M.gilvum_PYR-GCK -MTDRTKGRLELMTTRGMLTLEDLEKQVADGD----IDTVIVAFCDMQGR
MSMEG_2595|M.smegmatis_MC2_155 -----------MSRNPGMLTQDELVQLVTDGE----IDTVIVAFCDMQGR
Mvan_1603|M.vanbaalenii_PYR-1 MTNASIRPGRIDATNGQVESAPDAVRAMFERHG---IRTVQFGMADIDGQ
: * . : *: *
MMAR_3294|M.marinum_M LKSVAIAPAELEGAFEEG----------IGFDGSSIEGFAR----VSESD
MUL_1335|M.ulcerans_Agy99 LKSVAIAPAELEGAFEEG----------IGFDGSSIEGFAR----VSESD
MLBr_01631|M.leprae_Br4923 LKSVAIAPAELEGAFEEG----------IGFDGSSIEGFAR----VSESD
TH_3396|M.thermoresistible__bu LKSVAIAPAELEGAFEEG----------IGFDGSAIEGFAR----VSEAD
MAB_1920|M.abscessus_ATCC_1997 LKSVSIAPAELEGAFEEG----------IGFDGSSIEGFAR----VSESD
Mb2885c|M.bovis_AF2122/97 LAGKRISGRHFVDDIATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGD
Rv2860c|M.tuberculosis_H37Rv LAGKRISGRHFVDDIATRGVECCSYLLAVDVDLNTVPGYAMASWDTGYGD
MAV_3718|M.avium_104 LVGKRMDARLFVDEAAATGVECCGYLLAVDVDMNTVGGYAISGWDTGFGD
Mflv_4065|M.gilvum_PYR-GCK LTGKRISSRLFVDDVAEHGAECCNYLLAVDVDMNTVDGYAISSWEKGYGD
MSMEG_2595|M.smegmatis_MC2_155 LTGKRVSARLFVEEVAANGCECCNYLLAVDVDMNTVDGYAVSSWETGYGD
Mvan_1603|M.vanbaalenii_PYR-1 VRGKFVPAEFFLDSVARRGSSIPNIVFGWDIEDTLMDCFTVSGWHTGYSD
: . : : ..: . : :: . .*
MMAR_3294|M.marinum_M TVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLRRQ
MUL_1335|M.ulcerans_Agy99 TVAHPDPSTFQVLPWVTDAGHHHSARIFCDITMPDGSPSWADPRHVLRRQ
MLBr_01631|M.leprae_Br4923 TVAHPDPSTFQILPWATTAGHHHSARMFCDITMPDGSSSWADPRHVLRRQ
TH_3396|M.thermoresistible__bu MVARPDPSTFQVLPWTGDSGKHYSARMFCDITMPDGSPSWADPRHVLRRQ
MAB_1920|M.abscessus_ATCC_1997 TVAHPDPSTFQVLPWTTSAGKHHSARMFCDIKLPDGSPSWSDPRHVLRRQ
Mb2885c|M.bovis_AF2122/97 MVMTPDLSTLRLIPWLPG-----TALVIADLVWADGSEVAVSPRSILRRQ
Rv2860c|M.tuberculosis_H37Rv MVMTPDLSTLRLIPWLPG-----TALVIADLVWADGSEVAVSPRSILRRQ
MAV_3718|M.avium_104 LVMRPDLSTLRRIPWLPG-----TALVIADVVGADGSPVAVSPRAVLRRQ
Mflv_4065|M.gilvum_PYR-GCK MVMTPDMSTLRRVPWLPA-----TALVMADLSWTDGRPVTQAPRGILRAQ
MSMEG_2595|M.smegmatis_MC2_155 MVMTPDFSTLRRIPWLPG-----TAMVMADLSWLDGNPVAVAPRSILNHQ
Mvan_1603|M.vanbaalenii_PYR-1 VVLAPDLSTLRPVPWEPG-----HAFVLCDVVNTDGSPVPVAPRTVLAGQ
* ** **:: :** * ::.*: ** ** :* *
MMAR_3294|M.marinum_M LTKANELGFSCYVHPEIEFFLLNPGPEDG------TEPVPIDNAGYFDQA
MUL_1335|M.ulcerans_Agy99 LIKANELGFSCYVHPEIEFFLLNPGPEDG------TEPVPIDNAGYFDQA
MLBr_01631|M.leprae_Br4923 LTKANDLGFSCYVHPEIEFFLLKAGPEDGSM----PIPVPVDNAGYFDQA
TH_3396|M.thermoresistible__bu LAKAGDQGFTCYVHPEIEFFLLKPGPHDG------SPPVPADTSGYFDQS
MAB_1920|M.abscessus_ATCC_1997 LAKASDLGFSCYVHPEIEFFLLKNLPDDG------TAPIPADDAGYFDQA
Mb2885c|M.bovis_AF2122/97 LDRLKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAIL
Rv2860c|M.tuberculosis_H37Rv LDRLKARGLVADVATELEFIVFDQPYRQAWASGYRGLTPASDYNIDYAIL
MAV_3718|M.avium_104 LDRLAGRGLFADAATELEFMVFDEPYRQAWASGYRGLTPASDYNIDYAIS
Mflv_4065|M.gilvum_PYR-GCK LDRLDALGLDAVAATELEFMVFDTGFRDAWAAGYRNLKPATDYNIDYAMH
MSMEG_2595|M.smegmatis_MC2_155 IDRLAERGLVPFVGTELEFMVFDDSFRQAWSSGYHNLTPATDYNVDYAIH
Mvan_1603|M.vanbaalenii_PYR-1 VERAAALGYRLTAGYELEFYLYRETPETAAAKGYRNLTPASPGTATFSLN
: : * . *:** : . . :
MMAR_3294|M.marinum_M VHVSASKFRRHAIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADN
MUL_1335|M.ulcerans_Agy99 VHVSASKFRRHVIDALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADN
MLBr_01631|M.leprae_Br4923 VHDSALNFRRHAIEALEFMGISVEFSHHEGAPGQQEIDLRFADALSMADN
TH_3396|M.thermoresistible__bu VHDTAPNFRRHAIEALEQMGISVEFSHHEGAPGQQEIDLRYADALSMADN
MAB_1920|M.abscessus_ATCC_1997 VHEAAPNFRRHAIDALESMGISVEFSHHEGAPGQQEIDLRYADALSMADN
Mb2885c|M.bovis_AF2122/97 ASSRMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCDN
Rv2860c|M.tuberculosis_H37Rv ASSRMEPLLRDIRLGMAGAGLRFEAVKGECNMGQQEIGFRYDEALVTCDN
MAV_3718|M.avium_104 ASSRMEPLLRDIRRGMAGAGLRFESVKGECNRGQQEIGFRYDEALRTCDN
Mflv_4065|M.gilvum_PYR-GCK ASTRMEPLLRDIRLGMDGAGMYCEGVKGECNLGQQEIGFRYEQALVTCDN
MSMEG_2595|M.smegmatis_MC2_155 ASTRMEPLLRDIRRGMESAGMYCEGVKGECNLGQQEIAFRYDHARVTCDN
Mvan_1603|M.vanbaalenii_PYR-1 RHSALEDVIGAIREGMRACDIPVLASNTEYGAGQIEINIEHADALTTADR
. .: .: : * ** ** :.. .* .*.
MMAR_3294|M.marinum_M VMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEGDVN----
MUL_1335|M.ulcerans_Agy99 VMTFRYVIKEVALEEGVRATFMPKPFGQHAGSAMHTHMSLFEGDVN----
MLBr_01631|M.leprae_Br4923 VMTFRYVIKEVAIENGARASFMPKPFAQHPGSAMHTHMSLFEGDVN----
TH_3396|M.thermoresistible__bu VMTFRYVVKQVAISDGVRASFMPKPFAEHPGSAMHTHMSLFEGDTN----
MAB_1920|M.abscessus_ATCC_1997 VMTFRNVVKEVAIIDGVHATFMPKPFSDHPGSAMHTHMSLFEGDTN----
Mb2885c|M.bovis_AF2122/97 HAIYKNGAKEIADQHGKSLTFMAKYD-EREGNSCHIHVSLRGTDGS----
Rv2860c|M.tuberculosis_H37Rv HAIYKNGAKEIADQHGKSLTFMAKYD-EREGNSCHIHVSLRGTDGS----
MAV_3718|M.avium_104 HVIYKNGAKEIADQHGKSLTFMAKYD-EREGNSCHVHLSLRDAQGG----
Mflv_4065|M.gilvum_PYR-GCK HTIYKNGAKEIADQHGKSLTFMAKFD-ESEGNSCHIHISFRGRDGSPEDI
MSMEG_2595|M.smegmatis_MC2_155 HTIYRNGAKEIADQHGKSLTFMAKYD-EREGNSCHIHISLRGAGENGD--
Mvan_1603|M.vanbaalenii_PYR-1 VAIYKNGVRRIAEQHGYLATFMAKVATDSAGSSAHIHQSMQQLDAPAR--
:: :.:* .* :**.* : *.: * * *:
MMAR_3294|M.marinum_M -AFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGEA
MUL_1335|M.ulcerans_Agy99 -AFHSPDDPLQLSEVGKSFIAGILEHASEISAVTNQWVNSYKRLILGGEA
MLBr_01631|M.leprae_Br4923 -AFHSDDDPLQLSDVGKSFIAGILEHASEISAVTNQWVNSYKRLVVGGEA
TH_3396|M.thermoresistible__bu -AFHSPDDPLQLSDVAKSFIAGILEHANEISAVTNQWVNSYKRLVHGGEA
MAB_1920|M.abscessus_ATCC_1997 -AFHNPDDPLQLSDVGKSFIAGILEHANEISAVTNQWVNSYKRLVQGGEA
Mb2885c|M.bovis_AF2122/97 AVFADSNGPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSFA
Rv2860c|M.tuberculosis_H37Rv AVFADSNGPHGMSSMFRSFVAGQLATLREFTLCYAPTINSYKRFADSSFA
MAV_3718|M.avium_104 AAFADPSRPHGMSTMFCSFLAGLLATMADFTLFYAPNINSYKRFADESFA
Mflv_4065|M.gilvum_PYR-GCK AVFADDSDELGMSSMFRSFIAGQLATLREFTLFYAPNINSYKRFAEGSFA
MSMEG_2595|M.smegmatis_MC2_155 AVFADESDPHGMSPLFRSFIAGILATLRELTLFYAPNINSYKRFVDGSFA
Mvan_1603|M.vanbaalenii_PYR-1 NAMWDGDRP---SALMRQAVAGQLASMRDFTVLYCPTVNSYKRRVPGSWS
.: . . * : . :** * ::: :***** . :
MMAR_3294|M.marinum_M PTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLLA
MUL_1335|M.ulcerans_Agy99 PTAASWGAANRSALVRVPMYTPHKTSSRRIEVRSPDSACNPYLAFAVLLA
MLBr_01631|M.leprae_Br4923 PTTASWGAANRSALVRVPMYTPHKTSSRRVEVRSPDSACNPYLAFAVLLA
TH_3396|M.thermoresistible__bu PTAASWGAANRSALVRVPMYTPHKASSRRVEVRSPDSACNPYLTFAVLLA
MAB_1920|M.abscessus_ATCC_1997 PTAASWGAANRSALVRVPMYTPNKSSSRRIEVRSPDSACNPYLTYAVLLA
Mb2885c|M.bovis_AF2122/97 PTALAWGLDNRTCALRVVGHGQN----IRVECRVPGGDVNQYLAVAALIA
Rv2860c|M.tuberculosis_H37Rv PTALAWGLDNRTCALRVVGHGQN----IRVECRVPGGDVNQYLAVAALIA
MAV_3718|M.avium_104 PTALAWGLDNRTCALRVVGHGAH----TRVECRVPGGDVNPYLAVAAIVA
Mflv_4065|M.gilvum_PYR-GCK PTAVAWGLDNRTCSLRVVGHGQG----MRMENRAPGGDVNQYLAVAALIA
MSMEG_2595|M.smegmatis_MC2_155 PTAVAWGFDNRTCALRVVGHGHG----MRVECRAPGGDVNQYLAVSALIA
Mvan_1603|M.vanbaalenii_PYR-1 PVSAAWGVDNRTAALRVIAHDEAT---CRMENRVPGADANPYLALAACLA
*.: :** **:. :** : *:* * *.. * **: :. :*
MMAR_3294|M.marinum_M AGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMESS
MUL_1335|M.ulcerans_Agy99 AGLRGVEKGYVLGPQAEDNVWDLTPEERRTMGYRELPTSLDSALRAMESS
MLBr_01631|M.leprae_Br4923 AGLRGVEKGYVLGPQAEDNVWDLTPEERLAMGYRELPTSLDSALGAMEVS
TH_3396|M.thermoresistible__bu AGLRGVEKGYVLGPEAEDNVWTLTPEERRAMGFKELPSSLGQALAEMERS
MAB_1920|M.abscessus_ATCC_1997 AGLRGVEQGYTLGPEAEDDVWGLTRAERQAMGYKELPSTLENALIAMEGS
Mb2885c|M.bovis_AF2122/97 GGLYGIERGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFEDS
Rv2860c|M.tuberculosis_H37Rv GGLYGIERGLQLPEPCVGNAYQGADVER-------LPVTLADAAVLFEDS
MAV_3718|M.avium_104 GGLYGIEQGLALPEPCAGNAYRARGVGR-------LPATLAEAAALFEHS
Mflv_4065|M.gilvum_PYR-GCK GGLYGIENKLELEDAVEGNAYT-SGADR-------LPTTLSEAVELFENS
MSMEG_2595|M.smegmatis_MC2_155 GGLYGIERDLELPEPMAGNAYT-SGAER-------LPTTLAEAADLFAKS
Mvan_1603|M.vanbaalenii_PYR-1 GVLHGIENQLEPPAAVTGNAYQGADSP--------LPRSLAEAVDAFGAS
. * *:*. .:.: ** :* .* : *
MMAR_3294|M.marinum_M ELVAEALGEHVFDFFLRNKRREWATYRSHVTPYELSTYLSL-
MUL_1335|M.ulcerans_Agy99 ELVAEALGEHVFDFFLRNKRREWATYRSHVTPYELSTYLSL-
MLBr_01631|M.leprae_Br4923 EFVAEALGEHVFDFFLRNKRAEWANYRSHVSLYELKTYLSL-
TH_3396|M.thermoresistible__bu ELVAEALGEHVFDFFLRNKRAEWENYRSHVTPYELKNYLSL-
MAB_1920|M.abscessus_ATCC_1997 ELVAEALGEHVFDFFLRNKRAEWARYRSNVTPFELRAYLGL-
Mb2885c|M.bovis_AF2122/97 ALVREAFGEDVVAHYLNNARVELAAFNAAVTDWERIRGFERL
Rv2860c|M.tuberculosis_H37Rv ALVREAFGEDVVAHYLNNARVELAAFNAAVTDWERIRGFERL
MAV_3718|M.avium_104 ALARQVFGDDVVAHYLNNARVELAAFHAAVTDWERMRGFERL
Mflv_4065|M.gilvum_PYR-GCK KIARDAFGDDVVDHYVNNARVELKAFNAAVTDWERVRGFERL
MSMEG_2595|M.smegmatis_MC2_155 EVARAAFGDDVVEHYLNYARVEMAAFNAAVTDWERVRGFERL
Mvan_1603|M.vanbaalenii_PYR-1 SLARTAFGDAFVDHYLASRGWERDRHREAVSDWETGRYFSRV
.. .:*: .. .:: * .. *: :* :