For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLAQTATPPATSDRRIRSFGEIPSPHAVSTEFPLGARRAERVARDRDEIADILAGRDDRLLVVVGPCSV HDPAAALEYAGRLVKIAAELKDSLKIVMRVYFEKPRTTIGWKGLINDPGMDGTFDVARGLRIARQLLLDI IDIGLPVGCEFLEPTSPQYIADAVAWGAIGARTTESQVHRQLASGLSMPVGFKNGTDGNIQVAVDGAKSA AAQHVFFGMDDMGRGAVVSTEGNRDCHVILRGGTGGPNWDAESVRSAADKLESAGLPGRVVIDCSHANSG KDHVRQASVAAEVAQLVRDGLPVSGVMLESFLVAGAQAPEARPLTYGQSVTDKCMDWGATDLVLRELARR G
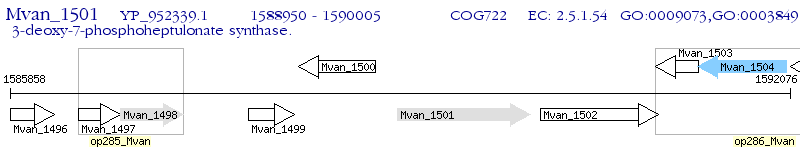
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1501 | - | - | 100% (351) | phospho-2-dehydro-3-deoxyheptonate aldolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4919 | - | 1e-177 | 87.18% (351) | phospho-2-dehydro-3-deoxyheptonate aldolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2007 | - | 1e-137 | 71.35% (349) | phospho-2-dehydro-3-deoxyheptonate aldolase |
| M. smegmatis MC2 155 | MSMEG_2799 | - | 0.0 | 89.43% (350) | phospho-2-dehydro-3-deoxyheptonate aldolase |
| M. thermoresistible (build 8) | TH_1686 | - | 1e-175 | 85.43% (350) | phospho-2-dehydro-3-deoxyheptonate aldolase |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1501|M.vanbaalenii_PYR-1 MTLAQTATPP-ATSDRRIRSFGEIPSPHAVSTEFPLGARRAERVARDRDE
MSMEG_2799|M.smegmatis_MC2_155 MNLAQIANPP-ATSDRRIRSFSAIPSPHDVLTEFPLGARRAERVAADRDE
Mflv_4919|M.gilvum_PYR-GCK MTLAQTATSQ-ETSDRRIRRFSEIPSPHDVLTEFPLGARRAERVARDREE
TH_1686|M.thermoresistible__bu MNSAQITTAP-KTSDRRIRGFLEIPSPHDVLSEFPLGARRAELVARDRDE
MAV_2007|M.avium_104 MSLTECATAAVDTSDHRIVSFRELSAPAAIRAELPLTPRRARAVQRDRDD
*. :: :.. ***:** * :.:* : :*:** .***. * **::
Mvan_1501|M.vanbaalenii_PYR-1 IADILAGRDDRLLVVVGPCSVHDPAAALEYAGRLVKIAAELKDSLKIVMR
MSMEG_2799|M.smegmatis_MC2_155 VADILAGRDNRLLVVVGPCSVHDPAAALEYASRLVKVAEELKDTVKIVMR
Mflv_4919|M.gilvum_PYR-GCK IADILAGRDDRLLVVVGPCSVHDPAAALDYASRLVKVADDLGDRLKVVMR
TH_1686|M.thermoresistible__bu IADILAGRDDRLLAVVGPCSVHDPAAALDYASRLVKVAEELRDDLKVVMR
MAV_2007|M.avium_104 IAAILAGADDRLLLVVGPCSVHDPVAALDYARRLAPIAAEFADRLKIVMR
:* **** *:*** **********.***:** **. :* :: * :*:***
Mvan_1501|M.vanbaalenii_PYR-1 VYFEKPRTTIGWKGLINDPGMDGTFDVARGLRIARQLLLDIIDIGLPVGC
MSMEG_2799|M.smegmatis_MC2_155 VYFEKPRTTIGWKGLINDPGMDGTFDVARGLRIARQLLLDIIDIGLPVGC
Mflv_4919|M.gilvum_PYR-GCK VYFEKPRTTTGWKGLINDPGMDGSFDVARGLRTARQLLLDIIDIGLPVGC
TH_1686|M.thermoresistible__bu VYFEKPRTTVGWKGLINDPAMDGTFDVARGLRIARQLLLDVIDIGLPVGC
MAV_2007|M.avium_104 VYFEKPRTTIGWKGLINDPAMDSSFDVERGIRTARALLLDIIDVGLPVGC
********* *********.**.:*** **:* ** ****:**:******
Mvan_1501|M.vanbaalenii_PYR-1 EFLEPTSPQYIADAVAWGAIGARTTESQVHRQLASGLSMPVGFKNGTDGN
MSMEG_2799|M.smegmatis_MC2_155 EFLEPTSPQYIADAVAWGAIGARTTESQVHRQLASGLSMPVGFKNGTDGN
Mflv_4919|M.gilvum_PYR-GCK EFLEPTSPQYIADAVAWGAIGARTTESQVHRQLASGLSMPVGFKNGTDGN
TH_1686|M.thermoresistible__bu EFLEPNSPQYIADAVAWGAIGARTTESQVHRQLASGLSMPVGFKNGTDGN
MAV_2007|M.avium_104 EFLEPTSPQYIADAVAWGAIGARTTESQVHRQLASGLSMPIGFKNGTDGN
*****.**********************************:*********
Mvan_1501|M.vanbaalenii_PYR-1 IQVAVDGAKSAAAQHVFFGMDDMGRGAVVSTEGNRDCHVILRGGTGGPNW
MSMEG_2799|M.smegmatis_MC2_155 VQVAVDGAKSAAAEHVFFGMDDMGRGAVVSTAGNEDCHVILRGGTRGPNY
Mflv_4919|M.gilvum_PYR-GCK IQVAVDGVKAAAAPHVFFGMTDMGRGALVSTTGNQDCHVILRGGTDGPNC
TH_1686|M.thermoresistible__bu IQVAVDGVKAAAAPHVFFGMDDLGRGALVSTEGNEDCHVILRGGSRGPNW
MAV_2007|M.avium_104 VQVAIDGVKAAAAPHHFFGTDDVGRAAVVETMGNPDCHVILRGGTAGPNY
:***:**.*:*** * *** *:**.*:*.* ** *********: ***
Mvan_1501|M.vanbaalenii_PYR-1 DAESVRSAADKLESAGLPGRVVIDCSHANSGKDHVRQASVAAEVAQLVRD
MSMEG_2799|M.smegmatis_MC2_155 DAESVRAAAAKLAKAGLPGRVVIDCSHANSGKDHIRQAVVATEVAQMVRD
Mflv_4919|M.gilvum_PYR-GCK DAASVTVTAEKLVTAGLPGRVVIDCSHANSGKDHIRQVQVAKDVAQLVRD
TH_1686|M.thermoresistible__bu DAESVRQTADMLTAAGLPGRVVVDCSHANSGKDHVRQAEVAVEVAKLVRA
MAV_2007|M.avium_104 DAGSVAGAIERLSAAGLSPRVMVDCSHANSAKDHVRQAEVAAELIARIAA
** ** : * ***. **::*******.***:**. ** :: :
Mvan_1501|M.vanbaalenii_PYR-1 GLP-VSGVMLESFLVAGAQAPEARPLTYGQSVTDKCMDWGATDLVLRELA
MSMEG_2799|M.smegmatis_MC2_155 GQP-ISGVMLESFLVGGAQSPDAETLTYGQSVTDKCMDWDATDLVLRELA
Mflv_4919|M.gilvum_PYR-GCK GLP-ISGVMLESFLVAGAQAPEAQPLTYGQSVTDKCMDWGATDLVLRELA
TH_1686|M.thermoresistible__bu GLP-ISGVMLESFLVAGAQSPDTKPLTYGQSVTDKCMDWATTDSVLRELA
MAV_2007|M.avium_104 GERGIGGLMLESFLVAGAQ-PLGGELTYGQSVTDECMDFATTARLLAGLY
* :.*:*******.*** * *********:***: :* :* *
Mvan_1501|M.vanbaalenii_PYR-1 RRG-
MSMEG_2799|M.smegmatis_MC2_155 RR--
Mflv_4919|M.gilvum_PYR-GCK NHG-
TH_1686|M.thermoresistible__bu KRR-
MAV_2007|M.avium_104 AAMG