For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSEDRKIPVRTPAGKTDGSIELPAELFDVEPNIALMHQVVNAQLAAKRQGTHSTKTRGEVSGGGKKPYR QKGTGRARQGSTRAPQFTGGGIVHGPKPRDYSERTPKKMIRAALRGALSDRARNDRIHAITEIVEGQTPS TKSAKSFLATLTDRRQVLVVIGRADEASALSVRNLPGVHVISPDQLNTYDVLKADDVVFSVEALNAYIAA NTKTEEVSA
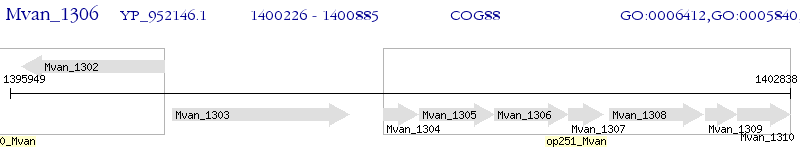
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1306 | rplD | - | 100% (219) | 50S ribosomal protein L4 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0722 | rplD | 1e-102 | 85.98% (214) | 50S ribosomal protein L4 |
| M. gilvum PYR-GCK | Mflv_5048 | rplD | 1e-104 | 87.96% (216) | 50S ribosomal protein L4 |
| M. tuberculosis H37Rv | Rv0702 | rplD | 1e-102 | 85.98% (214) | 50S ribosomal protein L4 |
| M. leprae Br4923 | MLBr_01862 | rplD | 2e-96 | 79.28% (222) | 50S ribosomal protein L4 |
| M. abscessus ATCC 19977 | MAB_3819c | rplD | 3e-94 | 81.69% (213) | 50S ribosomal protein L4 |
| M. marinum M | MMAR_1032 | rplD | 1e-100 | 82.87% (216) | 50S ribosomal protein L4, RplD |
| M. avium 104 | MAV_4470 | rplD | 1e-100 | 83.64% (214) | 50S ribosomal protein L4 |
| M. smegmatis MC2 155 | MSMEG_1437 | rplD | 1e-104 | 88.89% (207) | 50S ribosomal protein L4 |
| M. thermoresistible (build 8) | TH_0406 | rplD | 2e-99 | 82.65% (219) | PROBABLE 50S RIBOSOMAL PROTEIN L4 RPLD |
| M. ulcerans Agy99 | MUL_0791 | rplD | 4e-99 | 81.94% (216) | 50S ribosomal protein L4 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1032|M.marinum_M MAAQNQSAQKDLEIQVKTPDGKVDGSVALPAELFDVPANIALMHQVVTAQ
MUL_0791|M.ulcerans_Agy99 MAAQNQSAQKDLEIQVKAPDGKVDGSVVLPAELFDVPANIALMHQVVTAQ
MAB_3819c|M.abscessus_ATCC_199 -----------MKIAVKAPGGKTDGSIELPAELFDAPANIALLHQVVTAQ
MLBr_01862|M.leprae_Br4923 MAAKVENGSNIRKIDVKTPDGKVDGTIELPAELFDAPANIALMHQVVTAQ
Mb0722|M.bovis_AF2122/97 MAAQEQ---KTLKIDVKTPAGKVDGAIELPAELFDVPANIALMHQVVTAQ
Rv0702|M.tuberculosis_H37Rv MAAQEQ---KTLKIDVKTPAGKVDGAIELPAELFDVPANIALMHQVVTAQ
MAV_4470|M.avium_104 ---------MALKLDVKAPGGKVEGSIELPAELFDAPANIALMHQVVTAQ
Mvan_1306|M.vanbaalenii_PYR-1 ------MTSEDRKIPVRTPAGKTDGSIELPAELFDVEPNIALMHQVVNAQ
Mflv_5048|M.gilvum_PYR-GCK ------MAN--KTIDVVTPAGKKDGTVELPAALFDAEPNIALMHQVVTAQ
MSMEG_1437|M.smegmatis_MC2_155 ---------MTLKVDVKTPAGKTDGSVELPAELFDVEPNIALMHQVVTAQ
TH_0406|M.thermoresistible__bu -----------MKIDVRTPAGKTEGSVELPAELFDVEPNIALMHQVVTAQ
: * :* ** :*:: *** ***. .****:****.**
MMAR_1032|M.marinum_M RAAARQGTHSTKTRGDVSGGGRKPYRQKGTGRARQGSTRAPQFTGGGVVH
MUL_0791|M.ulcerans_Agy99 RAAARQGTHSTKTRGDVSGGGRKPYRQKGTGRARQGSTRTPQFTGGGVVH
MAB_3819c|M.abscessus_ATCC_199 LAAGRQGTHSTKTRGLVSGGGKKPYRQKGTGRARQGSTRAPQFTGGGIVH
MLBr_01862|M.leprae_Br4923 RAAARQGTHSTKTRGDVSGGGRKPYRQKGTGRARQGSMRAPQFTGGGIVH
Mb0722|M.bovis_AF2122/97 RAAARQGTHSTKTRGEVSGGGRKPYRQKGTGRARQGSTRAPQFTGGGVVH
Rv0702|M.tuberculosis_H37Rv RAAARQGTHSTKTRGEVSGGGRKPYRQKGTGRARQGSTRAPQFTGGGVVH
MAV_4470|M.avium_104 RAAARQGTHSTKTRGDVSGGGRKPYRQKGTGRARQGSTRAPQFTGGGVVH
Mvan_1306|M.vanbaalenii_PYR-1 LAAKRQGTHSTKTRGEVSGGGKKPYRQKGTGRARQGSTRAPQFTGGGIVH
Mflv_5048|M.gilvum_PYR-GCK LAAKRQGTHATKTRAMVSGGGKKPYRQKGTGRARQGSTRAPQFTGGGVVH
MSMEG_1437|M.smegmatis_MC2_155 LAAKRQGTHSTKTRGEVSGGGKKPYRQKGTGRARQGSTRAPQFTGGGTVH
TH_0406|M.thermoresistible__bu LAAARQGTHSTKTRGEVRGGGRKPWRQKGTGRARQGSLRAPQFTGGGVVH
** *****:****. * ***:**:************ *:******* **
MMAR_1032|M.marinum_M GPKPRDYSQRTPKKMIAAALRGALSDRARNGRIHAVTELVAGQTPSTKSA
MUL_0791|M.ulcerans_Agy99 GPKPRDYSQRTPKKMIAAALRGALSDRARNGRIHAVTELVAGQTPSTKSA
MAB_3819c|M.abscessus_ATCC_199 GPQPRDYSQRTPKKMIAAALRGALSDRARNGRIHAVTELVAGQEPSTKSA
MLBr_01862|M.leprae_Br4923 GPKLRDYSQRTPKKMIAAALRGALSDRARNGRIHAVTELVVGKTPSTKSA
Mb0722|M.bovis_AF2122/97 GPKPRDYSQRTPKKMIAAALRGALSDRARNGRIHAITELVEGQNPSTKSA
Rv0702|M.tuberculosis_H37Rv GPKPRDYSQRTPKKMIAAALRGALSDRARNGRIHAITELVEGQNPSTKSA
MAV_4470|M.avium_104 GPKPRDYSQRTPKKMIAAALRGALSDRARNGRIHAITELVSGQTPSTKSA
Mvan_1306|M.vanbaalenii_PYR-1 GPKPRDYSERTPKKMIRAALRGALSDRARNDRIHAITEIVEGQTPSTKSA
Mflv_5048|M.gilvum_PYR-GCK GPQPRDYSQRTPKKMIAAALRSALSDRARNDRIHAITEVVEGQSPSTKSA
MSMEG_1437|M.smegmatis_MC2_155 GPKPRDYSQRTPKKMIAAALRGALSDRARNDRIHAVTELVEGQTPSTKSA
TH_0406|M.thermoresistible__bu GPKPRDYSQRTPKKMIAAALRGALSDRARNGRIHAVTELVEGQTPSTKKA
**: ****:******* ****.********.****:**:* *: ****.*
MMAR_1032|M.marinum_M KTFLATITDRK-----QVLVVIGRDDQTGVKSVRNLPGVHILSPDQLNTY
MUL_0791|M.ulcerans_Agy99 KTFLATITDRK-----QVLVVIGRDDQTGVKSVRNLPGVHILSPDQLNTY
MAB_3819c|M.abscessus_ATCC_199 KSFLGEITDRK-----QLLVVLGRDDLASVKSVRNLPNVHVLAYDQLNTY
MLBr_01862|M.leprae_Br4923 KEFLGTLTDRK-----QVLVVIGRSDETGAKSVRNLPGVHLLSPDQLNTY
Mb0722|M.bovis_AF2122/97 RAFLASLTERK-----QVLVVIGRSDEAGAKSVRNLPGVHILAPDQLNTY
Rv0702|M.tuberculosis_H37Rv RAFLASLTERK-----QVLVVIGRSDEAGAKSVRNLPGVHILAPDQLNTY
MAV_4470|M.avium_104 KAFLGTLTERK-----QVLVVIGRSDEAGAKSVRNLPGVHILAPDQLNTY
Mvan_1306|M.vanbaalenii_PYR-1 KSFLATLTDRR-----QVLVVIGRADEASALSVRNLPGVHVISPDQLNTY
Mflv_5048|M.gilvum_PYR-GCK KAFLATLTDRK-----QVLVVIGRADETSALSVRNLPGVHVISPDQLNTY
MSMEG_1437|M.smegmatis_MC2_155 KTFLGTLTENK-----KVLVVIGRTDEVGAKSVRNLPGVHVISPDQLNTY
TH_0406|M.thermoresistible__bu KAFLSTVIADRTVEKDKVLVVIGRTDETSIRSVRNLPGVHVIAPDQLNTY
: **. : : ::***:** * .. ******.**::: ******
MMAR_1032|M.marinum_M DVLRADDVVFSVEALNAYIAAN------TSEEVSA
MUL_0791|M.ulcerans_Agy99 DVLRADDVVFSVEALNAYIAAN------TSEEVSA
MAB_3819c|M.abscessus_ATCC_199 DVLKADDVVFSVEALNGFINAK------KKEEVSA
MLBr_01862|M.leprae_Br4923 DVLRADDLVFSVEALNSYIAAQQAAGTSTPEKVSA
Mb0722|M.bovis_AF2122/97 DVLRADDVVFSVEALNAYIAAN----TTTSEEVSA
Rv0702|M.tuberculosis_H37Rv DVLRADDVVFSVEALNAYIAAN----TTTSEEVSA
MAV_4470|M.avium_104 DVLRADDVVFSVEALRAYIAAN----TGTPEEVSA
Mvan_1306|M.vanbaalenii_PYR-1 DVLKADDVVFSVEALNAYIAAN----TKT-EEVSA
Mflv_5048|M.gilvum_PYR-GCK DVLKADDVVFSVEALNAYIAAA----TKTTEEVSA
MSMEG_1437|M.smegmatis_MC2_155 DVLNADDVVFSVEALNAYISAN----SKEGASV--
TH_0406|M.thermoresistible__bu DVLKADDVVFSVEALNAYISAN----TSTAKEVSA
***.***:*******..:* * .*