For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSTDLSAKLFADACSVIPGGVNSPVRAFSSVGGTPRFITSANGCWLTDADGNRYVDLVCSWGPMILGHA HPAVVEAVQRAAADGLSFGAPTPSEAELAREIIDRVAPVELIRLVNSGTEATMSAIRLARGFTGRAKIVK FSGCYHGHSDALLADAGSGVATLGLPSSPGVTGAAAADTIVLPYNNVPAVEEVFGRFGDEIACVITEASP GNMGTVPPLPGFNAELRRITAAHGALLILDEVMTGFRVSRAGWYGVDPVEADLFTFGKVMSGGLPAAAFG GRAEVMNRLAPLGPVYQAGTLSGNPVAMAAGLATLRAADDAAYATLDVNADRLAGLIGDALTEAGVAHQI PRAGNMFSVFFGAEPVTDFAAARATETWRFPAFFHGLLDAGVYPPPSAYECWFVSTALDDDAFDKIAAAL PGAARAAAQATA
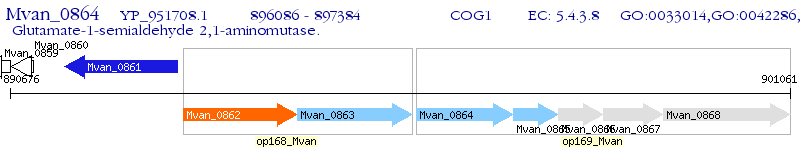
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0864 | - | - | 100% (432) | glutamate-1-semialdehyde aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_3311 | argD | 2e-32 | 33.65% (315) | acetylornithine aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_2918 | - | 2e-30 | 32.22% (329) | aminotransferase class-III |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0537 | hemL | 0.0 | 80.05% (441) | glutamate-1-semialdehyde aminotransferase |
| M. gilvum PYR-GCK | Mflv_0054 | - | 0.0 | 90.51% (432) | glutamate-1-semialdehyde aminotransferase |
| M. tuberculosis H37Rv | Rv0524 | hemL | 0.0 | 80.05% (441) | glutamate-1-semialdehyde aminotransferase |
| M. leprae Br4923 | MLBr_02414 | hemL | 0.0 | 79.91% (428) | glutamate-1-semialdehyde aminotransferase |
| M. abscessus ATCC 19977 | MAB_3978c | - | 0.0 | 77.99% (427) | glutamate-1-semialdehyde aminotransferase |
| M. marinum M | MMAR_0870 | hemL | 0.0 | 80.97% (431) | glutamate-1-semialdehyde 2,1-aminomutase, HemL |
| M. avium 104 | MAV_4621 | hemL | 0.0 | 83.96% (424) | glutamate-1-semialdehyde aminotransferase |
| M. smegmatis MC2 155 | MSMEG_0969 | hemL | 0.0 | 85.58% (430) | glutamate-1-semialdehyde aminotransferase |
| M. thermoresistible (build 8) | TH_4624 | - | 0.0 | 84.58% (428) | PUTATIVE glutamate-1-semialdehyde 2,1-aminomutase |
| M. ulcerans Agy99 | MUL_0623 | hemL | 0.0 | 81.21% (431) | glutamate-1-semialdehyde aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0864|M.vanbaalenii_PYR-1 ---------------MSSTDLSAKLFADACSVIPGGVNSPVRAFSSVGGT
Mflv_0054|M.gilvum_PYR-GCK ---------------MSTTEESAKLFADASSVIPGGVNSPVRAFNSVGGT
MSMEG_0969|M.smegmatis_MC2_155 MAMRVDQTCGSEEHSPASTQRSAQLFADACAVIPGGVNSPVRAFNSVGGT
Mb0537|M.bovis_AF2122/97 ----MGSTEQATSRVRGAARTSAQLFEAACSVIPGGVNSPVRAFTAVGGT
Rv0524|M.tuberculosis_H37Rv ----MGSTEQATSRVRGAARTSAQLFEAACSVIPGGVNSPVRAFTAVGGT
MLBr_02414|M.leprae_Br4923 ----MGSTDQATAPAGPAVSISAKLFEDACAVIPGGVNSPVRAFSAVGGT
MMAR_0870|M.marinum_M ----MGSTDQAVT----SVRRSAQLFTDACAVIPGGVNSPVRAFTAVGGT
MUL_0623|M.ulcerans_Agy99 ----MGSTDQAVT----SVRRSAQLFTDACAVIPGGVNSPVRAFTAVGGT
MAV_4621|M.avium_104 ----MGSSDQATAHAR-AQAASARLFDDACAVIPGGVNSPVRAFTAVGGT
TH_4624|M.thermoresistible__bu ------VVMRVDHPAPGTTRTSARLFADAATVIPGGVNSPVRAFAAVGGT
MAB_3978c|M.abscessus_ATCC_199 --MDDLASAETLDAVSDETPNSARLFADASTVIPGGVNSPVRAFAAVGGV
**:** *.:************* :***.
Mvan_0864|M.vanbaalenii_PYR-1 PRFITSANGCWLTDADGNRYVDLVCSWGPMILGHAHPAVVEAVQRAAADG
Mflv_0054|M.gilvum_PYR-GCK PRFITSAKGCRLTDVDGNQYVDLVCSWGPMILGHAHPAVVEAVQRVAAHG
MSMEG_0969|M.smegmatis_MC2_155 PRFITSAGGYWLTDADGNRYIDLVCSWGPMLLGHAHPAVVEAVRKVAEHG
Mb0537|M.bovis_AF2122/97 PRFITEAHGCWLIDADGNRYVDLVCSWGPMILGHAHPAVVEAVAKAAARG
Rv0524|M.tuberculosis_H37Rv PRFITEAHGCWLIDADGNRYVDLVCSWGPMILGHAHPAVVEAVAKAAARG
MLBr_02414|M.leprae_Br4923 PLFITEARGCWLTDADGRRYVDLVCSWGPMILGHAHPAVVDAVATVAASG
MMAR_0870|M.marinum_M PRFITEAHGCWLTDADGNRYADLVCSWGPMILGHAHPAVVEAVSRAATNG
MUL_0623|M.ulcerans_Agy99 PRFITEAHGCWLTDADGNRYADLVCSWGPMILGHAHPAVVEAVSRAAANG
MAV_4621|M.avium_104 PPFITAARGCRLTDADGNHYVDLVCSWGPMILGHAHPAVVEAVAKAAASG
TH_4624|M.thermoresistible__bu PRFITSARGCWLTDADGNRYVDLVCSWGPMILGHAHPAVVEAVQRAAADG
MAB_3978c|M.abscessus_ATCC_199 PRFVRSGAGCWLTDVDDNRYVDLVCSWGPLILGHAHPAVVEAVQRAAKDG
* *: . * * *.*..:* ********::*********:** .* *
Mvan_0864|M.vanbaalenii_PYR-1 LSFGAPTPSEAELAREIIDRVAPVELIRLVNSGTEATMSAIRLARGFTGR
Mflv_0054|M.gilvum_PYR-GCK LSFGAPTPSESELAREIIDRVAPVERVRLVNSGTEATMSAIRLARGFTGR
MSMEG_0969|M.smegmatis_MC2_155 LSFGAPTPSETELAAEIIDRVAPVEMMRFVNSGTEATMSAIRLARGFTGR
Mb0537|M.bovis_AF2122/97 LSFGAPTPAETQLAGEIIGRVAPVERIRLVNSGTEATMSAVRLARGFTGR
Rv0524|M.tuberculosis_H37Rv LSFGAPTPAETQLAGEIIGRVAPVERIRLVNSGTEATMSAVRLARGFTGR
MLBr_02414|M.leprae_Br4923 LSFGAPTPAETQLAAEIIGRMAPVERIRLVNSGTEATMSAVRLARGFTGR
MMAR_0870|M.marinum_M LSFGAPTPTETELAAEMIGRVAPVERLRLVNSGTEATMSAVRLARGFTGR
MUL_0623|M.ulcerans_Agy99 LSFGAPTPTETELAAEMIGRVAPVERLRLVNSGTEATMSAVRLARGFTGR
MAV_4621|M.avium_104 LSFGAPTPAESELAAEMIARVAPVERIRLVNSGTEATMSAVRLARGFTGR
TH_4624|M.thermoresistible__bu LSFGAPTPAETELVREIIDRVPPVQRLRLVNSGTEATMSAIRLARGFTGR
MAB_3978c|M.abscessus_ATCC_199 LSYGAPVEGEIELAREIIDRVQPVEKVRLVNSGTEATMSAVRLARGFTGR
**:***. * :*. *:* *: **: :*:***********:*********
Mvan_0864|M.vanbaalenii_PYR-1 AKIVKFSGCYHGHSDALLADAGSGVATLG------------------LPS
Mflv_0054|M.gilvum_PYR-GCK AKIVKFSGCYHGHSDALLADAGSGVATLG------------------LPS
MSMEG_0969|M.smegmatis_MC2_155 AKIVKFSGCYHGHSDALLADAGSGVATLG------------------LPS
Mb0537|M.bovis_AF2122/97 AKIVKFSGCYHGHVDALLADAGSGVATLGLCDDPQRPASPRSQSSRGLPS
Rv0524|M.tuberculosis_H37Rv AKIVKFSGCYHGHVDALLADAGSGVATLGLCDDPQRPASPRSQSSRGLPS
MLBr_02414|M.leprae_Br4923 TKIIKFSGCYHGHVDALLADAGSGVATL------------------SLPS
MMAR_0870|M.marinum_M SKIIKFSGCYHGHVDALLADAGSGVATLG------------------LPS
MUL_0623|M.ulcerans_Agy99 SKIIKFSGCYHGHVDALLADAGSGVATLG------------------LPS
MAV_4621|M.avium_104 AKIVKFSGCYHGHVDALLADAGSGVATLG------------------LPS
TH_4624|M.thermoresistible__bu AKIVKFAGCYHGHSDALLADAGSGVATLG------------------LPS
MAB_3978c|M.abscessus_ATCC_199 PKIVKFAGCYHGHADALLADAGSGLATFG------------------LPS
.**:**:****** **********:**: ***
Mvan_0864|M.vanbaalenii_PYR-1 SPGVTGAAAADTIVLPYNNVPAVEEVFGRFGDEIACVITEASPGNMGTVP
Mflv_0054|M.gilvum_PYR-GCK SPGVTGAAAADTLVLPYNNVAAVEEIFAQFGDEIACVITEASPGNMGTVP
MSMEG_0969|M.smegmatis_MC2_155 SPGVTGAAAADTIVIPYNNVAAVEDVFAHFGDEIACVISEASPGNMGTVP
Mb0537|M.bovis_AF2122/97 SPGVTGAAAADTIVLPYNDIDAVQQTFARFGEQIAAVITEASPGNMGVVP
Rv0524|M.tuberculosis_H37Rv SPGVTGAAAADTIVLPYNDIDAVQQTFARFGEQIAAVITEASPGNMGVVP
MLBr_02414|M.leprae_Br4923 SPGVTGAAAADTIVLPYNDIEAVRQTFARLGDQIAAVITEASPGNMGVVP
MMAR_0870|M.marinum_M SPGVTGAAAADTIVLPYNDLEAVRQTFAQHGEQIAAVITEASPGNMGVVP
MUL_0623|M.ulcerans_Agy99 SPGVTGAAAADTIVLPYNDLEAVRQTFAQHGEQIAAVITEASPGNMGVVP
MAV_4621|M.avium_104 SPGVTGATAADTIVLPYNDIDAVRQAFAEFGDQIAAVITEASPGNMGVVP
TH_4624|M.thermoresistible__bu SPGVTGAATADTIVLPYNDVTAVEQTFDRLGAEIAAVITEAAPGNMGAVP
MAB_3978c|M.abscessus_ATCC_199 SPGVTGASAADTIVLPYNDIGAVAQVFAEQGDHIAAVITEAAAGNMGTVA
*******::***:*:***:: ** : * . * .**.**:**:.****.*.
Mvan_0864|M.vanbaalenii_PYR-1 PLPGFNAELRRITAAHGALLILDEVMTGFRVSRAGWYGVDPVEADLFTFG
Mflv_0054|M.gilvum_PYR-GCK PLPGFNAALRRITAAHGALLIVDEVMTGFRVSRSGWYGLDPVDADLFTFG
MSMEG_0969|M.smegmatis_MC2_155 PLPGFNAALRRITAEHGALLILDEVMTGFRVSRSGWYGLDPVEADLLTFG
Mb0537|M.bovis_AF2122/97 PGPGFNAALRAITAEHGALLILDEVMTGFRVSRSGWYGIDPVPADLFAFG
Rv0524|M.tuberculosis_H37Rv PGPGFNAALRAITAEHGALLILDEVMTGFRVSRSGWYGIDPVPADLFAFG
MLBr_02414|M.leprae_Br4923 PAPGYNAALRAITAEHGALLIIDEVMTGFRVSRSGWYGLDPVAADLFIFG
MMAR_0870|M.marinum_M PGRGYNAALRAVTAEHGALLIVDEVMTGFRVSRSGWYGIDPVDADLFTFG
MUL_0623|M.ulcerans_Agy99 PGRGYNAALRAVTAEHGALLIVDEVMTGFRVSRSGWYGIDPVDADLFTFG
MAV_4621|M.avium_104 PAPGFNAALRALTAEHGALLIVDEVMTGFRVSRSGWYGIDPVDADLFTFG
TH_4624|M.thermoresistible__bu PQPGFNAALRRITAEHGALLISDEVMTGFRVSRSGWYGLESVEADLFTFG
MAB_3978c|M.abscessus_ATCC_199 PGPDFNAGLRRITENHGALLIMDEVMTGFRVSRSGWYGIDQVAGDLVTFG
* .:** ** :* ****** ***********:****:: * .**. **
Mvan_0864|M.vanbaalenii_PYR-1 KVMSGGLPAAAFGGRAEVMNRLAPLGPVYQAGTLSGNPVAMAAGLATLRA
Mflv_0054|M.gilvum_PYR-GCK KVMSGGLPAAAFGGTAEVMGRLAPLGPVYQAGTLSGNPVAMAAGLATLRA
MSMEG_0969|M.smegmatis_MC2_155 KVMSGGLPAAAFGGRTEVMSRLAPLGPVYQAGTLSGNPVAMAAGLTTLRH
Mb0537|M.bovis_AF2122/97 KVMSGGMPAAAFGGRAEVMQRLAPLGPVYQAGTLSGNPVAVAAGLATLRA
Rv0524|M.tuberculosis_H37Rv KVMSGGMPAAAFGGRAEVMQRLAPLGPVYQAGTLSGNPVAVAAGLATLRA
MLBr_02414|M.leprae_Br4923 KVMSGGMPAAAFGGRAEVMERLAPLGPVYQAGTLSGNPVAMAAGLATLRA
MMAR_0870|M.marinum_M KVMSGGLPAAAFGGRAEVMERLAPLGPVYQAGTLSGNPVAMAAGLATLRA
MUL_0623|M.ulcerans_Agy99 KVMSGGLPAAAFGGRAEVMERLAPLGPVYQAGTLSGNPVAMAAGLATLRT
MAV_4621|M.avium_104 KVMSGGLPAAAFGGRAEVMERLAPLGPVYQAGTLSGNPVAVAAGLATLRH
TH_4624|M.thermoresistible__bu KVMSGGLPAAAFGGRAEVMERLAPLGPVYQAGTLSGNPVAMAAGLATLRA
MAB_3978c|M.abscessus_ATCC_199 KVMSGGMPAAAFGGRADIMDRLAPLGPVYQAGTLSGNPVAVAAGLASLRA
******:******* :::* ********************:****::**
Mvan_0864|M.vanbaalenii_PYR-1 ADDAAYATLDVNADRLAGLIGDALTEAGVAHQIPRAGNMFSVFFGAEPVT
Mflv_0054|M.gilvum_PYR-GCK ADDAAYAKLDANADRLVGLIGDALTEAGVTHQIPRAGNMFSVFFGGDPVT
MSMEG_0969|M.smegmatis_MC2_155 ADDAVYAKLDANADRLAGLLTGALTEAGVAHQVQRAGNMLSVFFTDAPVH
Mb0537|M.bovis_AF2122/97 ADDAVYTALDANADRLAGLLSEALTDAVVPHQISRAGNMLSVFFGETPVT
Rv0524|M.tuberculosis_H37Rv ADDAVYTALDANADRLAGLLSEALTDAVVPHQISRAGNMLSVFFGETPVT
MLBr_02414|M.leprae_Br4923 AADAVYATLDRNADRLVAMLSEALTDAGVPHQIPRAGNMFSVFFSEAPVT
MMAR_0870|M.marinum_M ADDGVYAALDANADRLARLLAGALTDAGVAHQIPRAGNMLSVFFSETPVT
MUL_0623|M.ulcerans_Agy99 ADDGVYAALDANADRLARLLAGALTDAGVAHQIPRAGNMLSVFFSETPVT
MAV_4621|M.avium_104 ADAAAYAALDANADRLARLLSDALTNAGVPHQIPRAGNMLSVFFTDTPVT
TH_4624|M.thermoresistible__bu ADDAAYATLDANADRLTGLLTSALAEAGVPHQVSRAGNMLSVFFTDKPVT
MAB_3978c|M.abscessus_ATCC_199 ADAKAYETLDRNADALIALLTASLDAEGVAHHIGRAGNMFSVFFGTEPVR
* .* ** *** * :: :* *.*:: *****:**** **
Mvan_0864|M.vanbaalenii_PYR-1 DFAAARATETWRFPAFFHGLLDAGVYPPPSAYECWFVSTALDDDAFDKIA
Mflv_0054|M.gilvum_PYR-GCK DFASAKATETWRFPAFFHALLDAGIYPPPSAYEAWFVSTALDDEAFDRIA
MSMEG_0969|M.smegmatis_MC2_155 DFAAAKATETWRFPPFFHALLERGVYPPCSAFETWFVSAALDDDAFARIA
Mb0537|M.bovis_AF2122/97 DFASARASQTWRYPAFFHAMLDAGVYPPCSAFEAWFVSAALDDAAFGRIA
Rv0524|M.tuberculosis_H37Rv DFASARASQTWRYPAFFHAMLDAGVYPPCSAFEAWFVSAALDDAAFGRIA
MLBr_02414|M.leprae_Br4923 DFASACNSQVWRYPAFFHALLDAGVYPPCSTFEAWFVSAALDDAAFGRIV
MMAR_0870|M.marinum_M DFAAARNSQSWRFAPFFHALLDAGVYPPCSAFEAWFVSAALDDTAFERIA
MUL_0623|M.ulcerans_Agy99 DFAAARNSQTWRFAPFFHALLDAGVYPPCSAFEAWFVSAALDDTAFERIA
MAV_4621|M.avium_104 DFASARATQTWRYPPFFHALLDAGVYPPCSAFETWFVSTALDDDAFDRIA
TH_4624|M.thermoresistible__bu DYDTARASETWRFPPFFHALLERGVYAPPSAFEAWFVSTALDDEAFDKIA
MAB_3978c|M.abscessus_ATCC_199 DFQSAKATETWRYPAFFHGLLSRGVYPPASAFETWFVSTALDDEAFDRIA
*: :* :: **:..***.:*. *:*.* *::* ****:**** ** :*.
Mvan_0864|M.vanbaalenii_PYR-1 AALP--GAARAAAQATA-----
Mflv_0054|M.gilvum_PYR-GCK DALP--GAARAAAQAAA-----
MSMEG_0969|M.smegmatis_MC2_155 EALP--AAARAAAAATPEGTA-
Mb0537|M.bovis_AF2122/97 NALP--AAARAAAQERPA----
Rv0524|M.tuberculosis_H37Rv NALP--AAARAAAQERPA----
MLBr_02414|M.leprae_Br4923 DALPGAAAAAVAARHRES----
MMAR_0870|M.marinum_M DALP--TAARAAANATPESRP-
MUL_0623|M.ulcerans_Agy99 DALP--TAARAAANATPESRP-
MAV_4621|M.avium_104 AALP--AAARAAAGADEGNS--
TH_4624|M.thermoresistible__bu DALP--AAARAAGRAEPEARPA
MAB_3978c|M.abscessus_ATCC_199 DALP--AAARAAAAAEEN----
*** ** .*.