For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTHYDVVVLGAGPGGYVAAIRAAQLGLSTAIVEPKYWGGVCLNVGCIPSKALLRNAELAHIFTKEAKTFG ISGEASFDYGAAFDRSRKVADGRVAGVHFLMKKNKITEIHGYGKFTGPNSIEVDLNDGGTEKLEFDNAII ATGSSTRLVPGTSLSENVVTYEDQILSRELPGSIVIAGAGAIGMEFAYVMKNYGVDVTIVEFLPRALPNE DAEVSKEIEKQYKKLGVKILTGTKVEKIQDDGSTVTVTVSKDGKTDELKADKVLQAIGFAPNVEGFGLET TGVQLTDRRAIGIDDYMRTNVQHIYAIGDVTAKLQLAHVAEAMGVVAAETIAGAETLALGDYRMLPRATF CQPQVASFGLTEQQAKDEGYDVVVAKFPFTANGKAHGLGDPTGFVKLVADRKHLELLGGHLIGPDVSELL PELTLAQKWDLTANELARNVHTHPTLSEAMQECFHGLVGHMINF
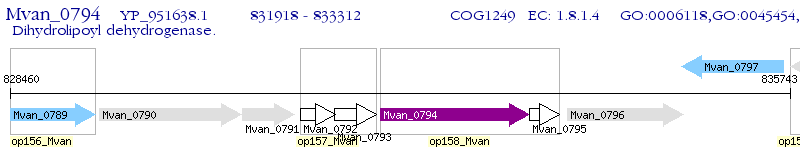
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0794 | - | - | 100% (464) | dihydrolipoamide dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3468 | - | 3e-49 | 28.16% (451) | mercuric reductase |
| M. vanbaalenii PYR-1 | Mvan_2448 | - | 1e-46 | 31.35% (453) | soluble pyridine nucleotide transhydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0471 | lpd | 0.0 | 85.78% (464) | dihydrolipoamide dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0114 | - | 0.0 | 91.22% (467) | dihydrolipoamide dehydrogenase |
| M. tuberculosis H37Rv | Rv0462 | lpd | 0.0 | 85.78% (464) | dihydrolipoamide dehydrogenase |
| M. leprae Br4923 | MLBr_02387 | lpd | 0.0 | 81.80% (467) | dihydrolipoamide dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4127c | - | 0.0 | 86.15% (462) | dihydrolipoamide dehydrogenase |
| M. marinum M | MMAR_0785 | lpd | 0.0 | 88.15% (464) | dihydrolipoamide dehydrogenase Lpd |
| M. avium 104 | MAV_4687 | lpdA | 0.0 | 87.47% (463) | dihydrolipoamide dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0903 | lpdA | 0.0 | 91.81% (464) | dihydrolipoamide dehydrogenase |
| M. thermoresistible (build 8) | TH_4200 | - | 0.0 | 83.26% (466) | PUTATIVE dihydrolipoamide dehydrogenase |
| M. ulcerans Agy99 | MUL_2214 | lpd | 0.0 | 87.72% (464) | dihydrolipoamide dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0794|M.vanbaalenii_PYR-1 ---MTHYDVVVLGAGPGGYVAAIRAAQLGLSTAIVEPKYWGGVCLNVGCI
Mflv_0114|M.gilvum_PYR-GCK ---MTHYDVVVLGAGPGGYVAAIRAAQLGLSTAIVEPKYWGGVCLNVGCI
MSMEG_0903|M.smegmatis_MC2_155 ---MTHFDVVVLGAGPGGYVAAIRAAQLGLNTAIVEPKYWGGVCLNVGCI
MAB_4127c|M.abscessus_ATCC_199 --MTAHYDVVVLGAGPGGYVAAIRAAQLGLKTAIVEEKYWGGVCLNVGCI
Mb0471|M.bovis_AF2122/97 ---MTHYDVVVLGAGPGGYVAAIRAAQLGLSTAIVEPKYWGGVCLNVGCI
Rv0462|M.tuberculosis_H37Rv ---MTHYDVVVLGAGPGGYVAAIRAAQLGLSTAIVEPKYWGGVCLNVGCI
MMAR_0785|M.marinum_M MGAVTHYDVVVLGAGPGGYVAAIRAAQLGLNTAIVEPKYWGGVCLNVGCI
MUL_2214|M.ulcerans_Agy99 ---MTHYDVVVLGAGPAGYVAAIRAAQLGLNTAIVEPKYWGGVCLNVGCI
MAV_4687|M.avium_104 --MTSHYDVVVLGAGPGGYVAAIRAAQLGLSTAIVEPKYWGGVCLNVGCI
MLBr_02387|M.leprae_Br4923 ---MTHYDVVVLGAGPGGYVAAIRAAQLGLSTAVVEPKYWGGICLNVGCI
TH_4200|M.thermoresistible__bu VEAVSHYDVVVLGAGPGGYVAAIRAAQLGQKTAIIEPRYWGGVCLNVGCI
:*:*********.************ .**::* :****:*******
Mvan_0794|M.vanbaalenii_PYR-1 PSKALLRNAELAHIFTKEAKTFGISGEASFDYGAAFDRSRKVADGRVAGV
Mflv_0114|M.gilvum_PYR-GCK PSKALLRNAELAHVFHKDAKTFGISGDVSFDYGAAFDRSRKVADGRVAGV
MSMEG_0903|M.smegmatis_MC2_155 PSKALLRNAEIAHMFNKEAKTFGISGEATFDYGAAFDRSRKVADGRVAGV
MAB_4127c|M.abscessus_ATCC_199 PSKALLRNAELAHIFTKEAKTFGISGEATFDFGAAFDRSRKVAEGRVAGV
Mb0471|M.bovis_AF2122/97 PSKALLRNAELVHIFTKDAKAFGISGEVTFDYGIAYDRSRKVAEGRVAGV
Rv0462|M.tuberculosis_H37Rv PSKALLRNAELVHIFTKDAKAFGISGEVTFDYGIAYDRSRKVAEGRVAGV
MMAR_0785|M.marinum_M PSKALLRNAELAHIFTKDAKAFGISGEATFDYGVAFDRSRKVAEGRVAGV
MUL_2214|M.ulcerans_Agy99 PSKALLRNAELAHIFTKDAKAFGISGEATFDYGVAFDRSRKVAEGRVAGV
MAV_4687|M.avium_104 PSKALLRNAELAHIFTKEAKTFGINGEATFDYGAAFDRSRKVAEGRVAGV
MLBr_02387|M.leprae_Br4923 PSKVLLHNAELAHIFTKEAKTFGISGDASFDYGIAYDRSRKVSEGRVAGV
TH_4200|M.thermoresistible__bu PSKALLRNAELASIFTRDAKLFGIQGEVTFDYGAAFDRSRKVADGRVSGV
***.**:***:. :* ::** ***.*:.:**:* *:******::***:**
Mvan_0794|M.vanbaalenii_PYR-1 HFLMKKNKITEIHGYGKFTGPNSIEVDLN-DG--GTEKLEFDNAIIATGS
Mflv_0114|M.gilvum_PYR-GCK HFLMKKNKITEVHGYGRFTGPNSIEVEPTGDGEEGPVKLEFDNAIIATGS
MSMEG_0903|M.smegmatis_MC2_155 HFLMKKNKITEIHGYGKFTGPNSLEVALN-DG--GTEKVEFSNAIIATGS
MAB_4127c|M.abscessus_ATCC_199 HFLMKKNKITEYEGVGTFTDANTLAVKKA-DG--SHETLTFANVIIATGS
Mb0471|M.bovis_AF2122/97 HFLMKKNKITEIHGYGTFADANTLLVDLNDGGTES---VTFDNAIIATGS
Rv0462|M.tuberculosis_H37Rv HFLMKKNKITEIHGYGTFADANTLLVDLNDGGTES---VTFDNAIIATGS
MMAR_0785|M.marinum_M HFLMKKNKITEIHGYGKFTDANTLSVDLNDGGTEK---VTFDNAIIATGS
MUL_2214|M.ulcerans_Agy99 HFLMKKNKITEIHGYGKFTDANTLSVDLNDGGTEK---VTFDNAIIATGS
MAV_4687|M.avium_104 HFLMKKNKITEIHGYGRFTDPHTLAVELNDGGTET---VTFDNAIIATGS
MLBr_02387|M.leprae_Br4923 HFLMKKNKITEIHGYGRFTDANTLSVELSEGVPETPLKVTFNNVIIATGS
TH_4200|M.thermoresistible__bu HFLMKKNKITEIHGYGRFTDAHTIEVELNEGGTEQ---VTFDNAIIATGS
*********** .* * *:..::: * . : * *.******
Mvan_0794|M.vanbaalenii_PYR-1 STRLVPGTSLSENVVTYEDQILSRELPGSIVIAGAGAIGMEFAYVMKNYG
Mflv_0114|M.gilvum_PYR-GCK STRLVPGTSLSENVVTYEKQILTRELPGSIIIAGAGAIGMEFAYVLKNYG
MSMEG_0903|M.smegmatis_MC2_155 STRLVPGTSLSENVVTYETQILSRELPGSIIIAGAGAIGMEFAYVLKNYG
MAB_4127c|M.abscessus_ATCC_199 SVRLVPGTSLSENVVTYEKQILTRELPGSIIIAGAGAIGMEFAYVLKNYG
Mb0471|M.bovis_AF2122/97 STRLVPGTSLSANVVTYEEQILSRELPKSIIIAGAGAIGMEFGYVLKNYG
Rv0462|M.tuberculosis_H37Rv STRLVPGTSLSANVVTYEEQILSRELPKSIIIAGAGAIGMEFGYVLKNYG
MMAR_0785|M.marinum_M STRLVPGTSLSTNVVTYEEQILTRELPKSIIIAGAGAIGMEFGYVLKNYG
MUL_2214|M.ulcerans_Agy99 STRLVPGTSLSTNVVTYEEQILTRELPKSIIIAGAGAIGMEFGYVLKNYG
MAV_4687|M.avium_104 STRLVPGTSLSANVVTYEEQILSRELPESIIIAGAGAIGMEFGYVLHNYG
MLBr_02387|M.leprae_Br4923 KTRLVPGTLLSTNVITYEEQILTRELPDSIVIVGAGAIGIEFGYVLKNYG
TH_4200|M.thermoresistible__bu STKLVPGTSLSENVVTYEKLIMTRELPGSIVIAGAGAIGMEFGYIMANYG
..:***** ** **:*** *::**** **:*.******:**.*:: ***
Mvan_0794|M.vanbaalenii_PYR-1 VDVTIVEFLPRALPNEDAEVSKEIEKQYKKLGVKILTGTKVEKIQDDGS-
Mflv_0114|M.gilvum_PYR-GCK VDVTIVEFLPRALPNEDAEVSKEIEKQYKKLGVKILTGTKVEKIEDDGK-
MSMEG_0903|M.smegmatis_MC2_155 VDVTIVEFLPRALPNEDAEVSKEIEKQYKKLGVKILTGTKVEGIADDGS-
MAB_4127c|M.abscessus_ATCC_199 VDVTIVEFLPRALPNEDAEVSKEIEKQYKKLGVKILTGTKVESITDSGS-
Mb0471|M.bovis_AF2122/97 VDVTIVEFLPRALPNEDADVSKEIEKQFKKLGVTILTATKVESIADGGS-
Rv0462|M.tuberculosis_H37Rv VDVTIVEFLPRALPNEDADVSKEIEKQFKKLGVTILTATKVESIADGGS-
MMAR_0785|M.marinum_M VDVTIVEFLPRALPNEDAEVSKEIERQFKKLGVKIRTGTKVESISDDGS-
MUL_2214|M.ulcerans_Agy99 VDVTIVEFLPRALPNEDAEVSKEIERQFKKLGVKIRTGTKVESISDDGS-
MAV_4687|M.avium_104 VEVTIVEFLPRALPNEDADVSKEIEKQFKKLGVKILTGTKVESISDDGS-
MLBr_02387|M.leprae_Br4923 VDVTIVEFLPRAMPNEDAEVSKEIEKQFKKMGIKILTGTKVESISDNGS-
TH_4200|M.thermoresistible__bu VDVTVVEFLPRVLPNEDPEVSREIEKQFKKLGVKIHTGTKVESITDNGAG
*:**:******.:****.:**:***:*:**:*:.* *.**** * *.*
Mvan_0794|M.vanbaalenii_PYR-1 -TVTVTVSKDGKTDELKADKVLQAIGFAPNVEGFGLETTGVQLTDRRAIG
Mflv_0114|M.gilvum_PYR-GCK -QVTVTVSKNDKTEELKADKVMQAIGFAPNVEGFGLDKAGVELTDRKAIG
MSMEG_0903|M.smegmatis_MC2_155 -TVKVTVSKDGKTEELKADKVLQAIGFAPNVEGYGLEAAGVELTDRKAIG
MAB_4127c|M.abscessus_ATCC_199 -QVTVKVSKDGQESELVADKVLQAIGFAPNVEGFGLENTGVALTDRGAIA
Mb0471|M.bovis_AF2122/97 -QVTVTVTKDGVAQELKAEKVLQAIGFAPNVEGYGLDKAGVALTDRKAIG
Rv0462|M.tuberculosis_H37Rv -QVTVTVTKDGVAQELKAEKVLQAIGFAPNVEGYGLDKAGVALTDRKAIG
MMAR_0785|M.marinum_M -AVTVVVSKDGKSEELKADKVLQAIGFAPNVEGYGLDKAGVALTDRKAIG
MUL_2214|M.ulcerans_Agy99 -AVTVVVSKGGKSEELKADKVLQAIGFAPNVEGYGLDKAGVALTDRKAIG
MAV_4687|M.avium_104 -QVTVVVSKDGNSQELKAAKVLQAIGFAPNVEGYGLEAAGVALTDRKAIG
MLBr_02387|M.leprae_Br4923 -HVLVAVSKDGQFQELKADKVLQAIGFAPNVDGYGLDKVGVALTADKAVD
TH_4200|M.thermoresistible__bu GSVVVKVSKDGESSELKADKVLQAIGFAPNVDGYGLDRAGVRLDDRQAIA
* * *:*.. .** * **:*********:*:**: .** * *:
Mvan_0794|M.vanbaalenii_PYR-1 IDDYMRTNVQHIYAIGDVTAKLQLAHVAEAMGVVAAETIAGAETLALGDY
Mflv_0114|M.gilvum_PYR-GCK IDDYMRTNVPHIYAIGDVTAKLQLAHVAEAMGVVAAETIAGADTLPFEDY
MSMEG_0903|M.smegmatis_MC2_155 IDDYMRTNVGHIYAIGDVTGKLQLAHVAEAMAVVAAETIAGAETLPLGDY
MAB_4127c|M.abscessus_ATCC_199 IDERMRTNVPHIYAIGDVTSKLQLAHVAEAQAVVAAETIAGAETLELGDY
Mb0471|M.bovis_AF2122/97 VDDYMRTNVGHIYAIGDVNGLLQLAHVAEAQGVVAAETIAGAETLTLGDH
Rv0462|M.tuberculosis_H37Rv VDDYMRTNVGHIYAIGDVNGLLQLAHVAEAQGVVAAETIAGAETLTLGDH
MMAR_0785|M.marinum_M IGEYMQTSVGHIYAIGDVTGQLMLAHVAEAMGVVAAETIAGAETLSLGDY
MUL_2214|M.ulcerans_Agy99 IGEYMQTSVGHIYAIGDVTGQLMLAHVAEAMGVVAAETIAGAETLPLGDY
MAV_4687|M.avium_104 ITDYMRTNIEHIYAIGDVTGKLQLAHVAEAQGVVAAETIAGAETLALGDY
MLBr_02387|M.leprae_Br4923 IDDYMQTNVSHIYAIGDVTGKLQLAHVAEAQGVVAAEAIAGAETLALSDY
TH_4200|M.thermoresistible__bu IDDYMRTSVPHIYAIGDVTAKLQLAHVAEAMGVVAAETIAGVETLPLGDY
: : *:*.: ********.. * ******* .*****:***.:** : *:
Mvan_0794|M.vanbaalenii_PYR-1 RMLPRATFCQPQVASFGLTEQQAKDEGYDVVVAKFPFTANGKAHGLGDPT
Mflv_0114|M.gilvum_PYR-GCK RMMPRATFCQPQVASFGLTEQQAKDEGYDVVVAKFPFTANGKAHGLGDPT
MSMEG_0903|M.smegmatis_MC2_155 RMMPRATFCQPQVASFGLTEEQARDEGYDVKVAKFPFTANGKAHGLADPT
MAB_4127c|M.abscessus_ATCC_199 RMMPRATFCQPQVASFGLTEEQARAEGYDVKVAKFPFTANGKAHGLADPT
Mb0471|M.bovis_AF2122/97 RMLPRATFCQPNVASFGLTEQQARNEGYDVVVAKFPFTANAKAHGVGDPS
Rv0462|M.tuberculosis_H37Rv RMLPRATFCQPNVASFGLTEQQARNEGYDVVVAKFPFTANAKAHGVGDPS
MMAR_0785|M.marinum_M RMLPRATFCQPQVASFGLTEQQARDEGYDVKVAKFPFTANGKAHGLGDPS
MUL_2214|M.ulcerans_Agy99 RMLPRATFCQPQVASFGLTEQQARDEGYDVKVAKFPFTANGKAHGLGDPS
MAV_4687|M.avium_104 RMMPRATFCQPNVASFGLTEQQARDEGHDVVVAKFPFTANGKAHGVGDPS
MLBr_02387|M.leprae_Br4923 RMMPRATFCQPNVASFGLTEQQARDGGYDVVVAKFPFTANAKAHGMGDPS
TH_4200|M.thermoresistible__bu RMMPRATFCQPQVASFGLTEEQAREEGYDVKVAKFPFTANGKAHGMGAPS
**:********:********:**: *:** *********.****:. *:
Mvan_0794|M.vanbaalenii_PYR-1 GFVKLVADRKHLELLGGHLIGPDVSELLPELTLAQKWDLTANELARNVHT
Mflv_0114|M.gilvum_PYR-GCK GFVKLIADKKHLELLGGHLIGPDVAELLPELTLAQKWDLTANELARNVHT
MSMEG_0903|M.smegmatis_MC2_155 GFVKLIADAKYGELLGGHLIGPDVSELLPELTLAQKWDLTVNELARNVHT
MAB_4127c|M.abscessus_ATCC_199 GFVKLIADAKYGELIGGHLIGPDVSELLPELTLAQKWDLTVNELTRNVHT
Mb0471|M.bovis_AF2122/97 GFVKLVADAKHGELLGGHLVGHDVAELLPELTLAQRWDLTASELARNVHT
Rv0462|M.tuberculosis_H37Rv GFVKLVADAKHGELLGGHLVGHDVAELLPELTLAQRWDLTASELARNVHT
MMAR_0785|M.marinum_M GFVKLVADGKHGELLGGHLVGHDVSELLPELTLAQKWDLTATELARNVHT
MUL_2214|M.ulcerans_Agy99 GFVKLVADGKHGELLGGHLVGHDVSELLPELTLAQKWDLTATELARNVHT
MAV_4687|M.avium_104 GFVKLIADAKYGELLGGHLVGHDVSELLPELTLAQKWDLTATELARNVHT
MLBr_02387|M.leprae_Br4923 GFVKLVADAKYGELLGGHMIGHNVSELLPELTLAQKWDLTATELVRNVHT
TH_4200|M.thermoresistible__bu GFVKLIADAKYGELLGGHMVGHDVSELLPELTLAQKWDLTAHELARNVHT
*****:** *: **:***::* :*:**********:****. **.*****
Mvan_0794|M.vanbaalenii_PYR-1 HPTLSEAMQECFHGLVGHMINF
Mflv_0114|M.gilvum_PYR-GCK HPTLSEAMQECFHGLVGHMINF
MSMEG_0903|M.smegmatis_MC2_155 HPTLSEALQECFHGLAGHMINF
MAB_4127c|M.abscessus_ATCC_199 HPTLSEALQEAFHGLAGHMINF
Mb0471|M.bovis_AF2122/97 HPTMSEALQECFHGLVGHMINF
Rv0462|M.tuberculosis_H37Rv HPTMSEALQECFHGLVGHMINF
MMAR_0785|M.marinum_M HPTMSEALQECFHGLAGHMINF
MUL_2214|M.ulcerans_Agy99 HPTMSEALQECFHGLAGHMINF
MAV_4687|M.avium_104 HPTMSEALQECFHGLTGHMINF
MLBr_02387|M.leprae_Br4923 HPTLSEALQECFHGLIGHMINF
TH_4200|M.thermoresistible__bu HPTLSEAMQECFHGLVGHMINF
***:***:**.**** ******