For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VPAHTRGTLAVIGGGVIGLSVARRALQDGWAVRVHRAERAGASWVAGGMLAPHSEGWPGEERLLQIGLES LRLWREGFLDGLPPEVVTARESLVVAVDPADAADLKTVAAWLSGQGHPVTLTTSAREVEPLLAQGIRHGF RAETELAVDNRAVVAALAAECERLGVEWAPPVAALTDAGPADTVVIANGIDAPTLWPGLPVRPVKGEVLR LRWRRGCLPVPQRVVRARVHGRQVYVVPRADGVVVGATQYEHGRDTAPTVSGVRDLLDDACAVMPALGEY EFAEAAAGLRPMTPDNLPLVGRLDERTLVAAGHGRSGFLLAPWTAESIAAELEGALVR
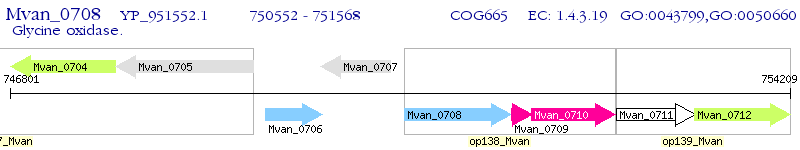
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0708 | - | - | 100% (338) | glycine oxidase ThiO |
| M. vanbaalenii PYR-1 | Mvan_4780 | - | 6e-05 | 27.61% (163) | D-amino-acid dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0423 | thiO | 1e-151 | 79.70% (330) | thiamine biosynthesis oxidoreductase ThiO |
| M. gilvum PYR-GCK | Mflv_0197 | - | 1e-168 | 85.76% (337) | glycine oxidase ThiO |
| M. tuberculosis H37Rv | Rv0415 | thiO | 1e-152 | 80.00% (330) | thiamine biosynthesis oxidoreductase ThiO |
| M. leprae Br4923 | MLBr_00299 | - | 1e-143 | 77.37% (327) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4218c | - | 1e-125 | 65.87% (334) | thiamine biosynthesis oxidoreductase ThiO |
| M. marinum M | MMAR_0718 | thiO | 1e-154 | 82.57% (327) | thiamine biosynthesis oxidoreductase ThiO |
| M. avium 104 | MAV_4746 | thiO | 1e-156 | 82.87% (327) | glycine oxidase ThiO |
| M. smegmatis MC2 155 | MSMEG_0791 | thiO | 1e-154 | 81.90% (326) | glycine oxidase ThiO |
| M. thermoresistible (build 8) | TH_2312 | - | 1e-159 | 82.01% (328) | POSSIBLE THIAMINE BIOSYNTHESIS OXIDOREDUCTASE THIO |
| M. ulcerans Agy99 | MUL_2805 | thiO | 1e-154 | 82.57% (327) | thiamine biosynthesis oxidoreductase ThiO |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0423|M.bovis_AF2122/97 -MASDLHTGSLAVIGGGVIGLSVARRAAQAGWPVRVHRSDERGASWVAGG
Rv0415|M.tuberculosis_H37Rv -MASDLHTGSLAVIGGGVIGLSVARRAAQAGWPVRVHRSDERGASWVAGG
MLBr_00299|M.leprae_Br4923 -MPSGPPAGSLAVIGGGVIGLSVARRAVQAGWSVRVHRTHEQGASWVAGG
MMAR_0718|M.marinum_M -MSS-PSMGSLAVIGGGVIGLSVARRAAQAGWAVRVHRRAEPGASWAAGG
MUL_2805|M.ulcerans_Agy99 -MSS-PSMGSLAVIGGGVIGLSVARRAAQAGWAVRVHRRAEPGASWAAGG
MAV_4746|M.avium_104 -MPS--DCGSLAVVGGGVIGLAVARRAAQAGWSVRVHRSGERGASWVAGG
Mvan_0708|M.vanbaalenii_PYR-1 --MPAHTRGTLAVIGGGVIGLSVARRALQDGWAVRVHRAERAGASWVAGG
Mflv_0197|M.gilvum_PYR-GCK --MPAHTRGRLAVIGGGVIGLAVARRAALDGWATRVHRTEHPGASWVAGG
MSMEG_0791|M.smegmatis_MC2_155 -----MTR-TVAVIGGGVIGLSVARRAALDGWTVRLHCTTERGASWVAGG
TH_2312|M.thermoresistible__bu MSDVIGSDRTVAVVGGGVIGLAVARRAALAGWQVRVHRSEERGASWVAGG
MAB_4218c|M.abscessus_ATCC_199 --MMSSQKGSVAVIGGGVIGLSVAREAARTGWRVVIHDDGAKGPSWVAGG
:**:*******:***.* ** . :* *.**.***
Mb0423|M.bovis_AF2122/97 MLAPHSEGWPGEERLLRLGLQSLRLWREGSFLDGLGPQLVTAHESLVVAV
Rv0415|M.tuberculosis_H37Rv MLAPHSEGWPGEERLLRLGLQSLRLWREGSFLDGLGPQLVTAHESLVVAV
MLBr_00299|M.leprae_Br4923 MLAPHSEGRPGEEQLLQLGLRSLRLWHEGGFTDRLAPEVITARESLVVAV
MMAR_0718|M.marinum_M MLAPHSEGWPGEERLLRLGLESLRLWHESGFLDGLPPQVVTARESLVVAV
MUL_2805|M.ulcerans_Agy99 MLAPHSEGWPGEERLLRLGLESLRLWHESGFLDGLPPQVVTARESLMVAV
MAV_4746|M.avium_104 MLAPHSEGWPGEERLLRLGLESLRLWREGGFLDGLPGRLVTARESLVVAV
Mvan_0708|M.vanbaalenii_PYR-1 MLAPHSEGWPGEERLLQIGLESLRLWREG-FLDGLPPEVVTARESLVVAV
Mflv_0197|M.gilvum_PYR-GCK MLAPHSEGWPGEEQLLTLGLESLRLWHEQ-FLGSLPAQVVTARESLVVAV
MSMEG_0791|M.smegmatis_MC2_155 MLAPHSEGWPGEEQLLQLGLESLRLWHSG-FLDGLPEKVVTARESLVVAV
TH_2312|M.thermoresistible__bu MLAPHSEGWPGEEHLLRLGLASLNLWRNG-FLDGLPPEVVTARESLVVAV
MAB_4218c|M.abscessus_ATCC_199 MLAPYSEAWPGEDELLELGIESLGIWRDG-FIDDVENPVITARGSLTVAV
****:**. ***:.** :*: ** :*:. * . : ::**: ** ***
Mb0423|M.bovis_AF2122/97 DRADVADLRTVADWLSAQGHPVIWESAARDVEPLLAQGIRHGFRAPTELA
Rv0415|M.tuberculosis_H37Rv DRADVADLRTVADWLSAQGHPVIWESAARDVEPLLAQGIRHGFRAPTELA
MLBr_00299|M.leprae_Br4923 DQADVADLRTVADWLSTQGHPVIWESAARDIEPLLAQGIRHGFWAPNELA
MMAR_0718|M.marinum_M DRADVADLRTVADWLSEQGHPVSWESAARDVEPLLAQGIRHGFRATTELA
MUL_2805|M.ulcerans_Agy99 DRADVADLRTVADWLSEQGHPVSWESAARDVEPLLAQGIRHGFRATTELA
MAV_4746|M.avium_104 DRADVADLRTVADWLSAQGHPVVWESAARDVEPLLAQGIRHGFTAPTELA
Mvan_0708|M.vanbaalenii_PYR-1 DPADAADLKTVAAWLSGQGHPVTLTTSAREVEPLLAQGIRHGFRAETELA
Mflv_0197|M.gilvum_PYR-GCK DQADAADLRTVAAWLCAQGHPVTPTTSARDMEPLLAQGIRHGFSADTELA
MSMEG_0791|M.smegmatis_MC2_155 DRADAADLRTVAEWLAAQGHPVEFTTAARDVEPLLAQGIRHGFRATTELA
TH_2312|M.thermoresistible__bu DRADVADLQTVGSWLAGQGHPVTFTTSARDVEPLLAQGIRHGFIADTELA
MAB_4218c|M.abscessus_ATCC_199 DTADVGELRTMTGWLAARGHPVTEIANARDVEPMLAQNVRRGFTAPEELS
* **..:*:*: **. :**** : **::**:***.:*:** * **:
Mb0423|M.bovis_AF2122/97 VDNRALLDVLCRDCERLGVRWSSQVSSLSDV-DAHTVVIANGIDAPALWP
Rv0415|M.tuberculosis_H37Rv VDNRALLDALCRDCERLGVRWSSQVSSLSDV-DAHTVVIANGIDAPALWP
MLBr_00299|M.leprae_Br4923 VDNRALLAGLCLDCERLGVRWASPVNDLSCV-NADAVVIANGIGAPALWP
MMAR_0718|M.marinum_M VDNRALLDALAVDCERRGVTWAGPVADLSDA-TGDAVVIANGIDAPALWP
MUL_2805|M.ulcerans_Agy99 VDNRALLDALAVDCERRGVTWAGPVADLSDA-TGDAVVIANGIDAPALWP
MAV_4746|M.avium_104 VDNRAVLEALDAACERLGVRWAPPLHDLAEA-DADAVVIANGIDAPALWP
Mvan_0708|M.vanbaalenii_PYR-1 VDNRAVVAALAAECERLGVEWAPPVAALTDAGPADTVVIANGIDAPTLWP
Mflv_0197|M.gilvum_PYR-GCK VDNRAVVAALAEQCERLDVRWAPPVDSLAETADADAVVIANGIDAPALWP
MSMEG_0791|M.smegmatis_MC2_155 VDNRGVVDALAAHCERLAVRWAGQVDDLADV-DADAVVIANGIDAPRLWP
TH_2312|M.thermoresistible__bu VDNRAVVEALRAHCDRLGVQWAPPVTDLAEV-DAPVRVIANGLDAPALWP
MAB_4218c|M.abscessus_ATCC_199 VDNRKVLQALTAECEALGVGRGPAAVHTRDV-DADQVVIANGVQAAALA-
**** :: * *: * . . . *****: *. *
Mb0423|M.bovis_AF2122/97 GLPIRPVKGEVLRLRWRPGCMPLPQRVIRARVRGRQVYLVPRSDGVVVGA
Rv0415|M.tuberculosis_H37Rv GLPIRPVKGEVLRLRWRPGCMPLPQRVIRARVRGRQVYLVPRSDGVVVGA
MLBr_00299|M.leprae_Br4923 GLPVRPVKGEVLRLRWRNGCMSLLKRLIRAHVHGRQVYLVPRADGVVVGA
MMAR_0718|M.marinum_M GLPVRPVKGEVLRLRWRKGCMPLLQRVVRARVHGRQVYLVPRADGVVVGA
MUL_2805|M.ulcerans_Agy99 GLPVRPVKGEVLRLRWRKGCMPLLQRVVRARVHGRQVYLVPRADGVVVGA
MAV_4746|M.avium_104 GLPVRPVKGEVLRLRWRKGCMPVPQRVIRARVHGRQVYLVPRADGVVVGA
Mvan_0708|M.vanbaalenii_PYR-1 GLPVRPVKGEVLRLRWRRGCLPVPQRVVRARVHGRQVYVVPRADGVVVGA
Mflv_0197|M.gilvum_PYR-GCK GLPVRPVKGEVLRLRWRRGCLPVPRRVVRARVRGRQVYVVPRPDGVVVGA
MSMEG_0791|M.smegmatis_MC2_155 DLPVRPVKGEVLRLRWRKGCMPVPQRVIRARVHGRQVYLVPRADGVVVGA
TH_2312|M.thermoresistible__bu GLPIRPVKGEVLRLRWRRGCMPVPQRVIRARVHGRQVYLVPRADGLVVGA
MAB_4218c|M.abscessus_ATCC_199 GLPVRPVKGEVLRLRQRPGAMPPPANVIRARVHGRHVYLVPRADGVVVGA
.**:*********** * *.:. .::**:*:**:**:***.**:****
Mb0423|M.bovis_AF2122/97 TQYEHGRDTAPVVSGVRDLLDDACTVLPALGEYELAECEAGLRPMTPDNL
Rv0415|M.tuberculosis_H37Rv TQYEHGRDTAPVVSGVRDLLDDACTVLPALGEYELAECEAGLRPMTPDNL
MLBr_00299|M.leprae_Br4923 TQYEHGYDTAPVVSGVRDLLDDACAVLPTLGEYELAECAAGLRPTTPNNL
MMAR_0718|M.marinum_M TQYEHGRDLAPVVSGVRDLLEDACAVMPALGEYELAECTAGLRPMTPDNL
MUL_2805|M.ulcerans_Agy99 TQYEHGRDLAPVVSGVRDLLEDACAVMPALGEYEFAECTAGLRPMTPDNL
MAV_4746|M.avium_104 TQYEHGRDTAPVVSGVRDLLDDACAVLPALGEYELAECGAGLRPMTPDNL
Mvan_0708|M.vanbaalenii_PYR-1 TQYEHGRDTAPTVSGVRDLLDDACAVMPALGEYEFAEAAAGLRPMTPDNL
Mflv_0197|M.gilvum_PYR-GCK TQYEHGRDTAPTVGGVRDLLDDACAVMPGLGEYEFAEAAAGLRPMTPDNL
MSMEG_0791|M.smegmatis_MC2_155 TQYEHGRDTAPVVSGVRDLLEDACAVMPALGEYELAEIAAGLRPMTPDGL
TH_2312|M.thermoresistible__bu TQYEHGRDTAPSVTGVRDLLDDSCAVVPALGEYELAECAAGLRPMTPDNL
MAB_4218c|M.abscessus_ATCC_199 TQYEHGHDTAPAVAGVRDLLDDACALLPCLGEYEFAECAAGLRPMTDDNL
****** * ** * ******:*:*:::* *****:** ***** * :.*
Mb0423|M.bovis_AF2122/97 PLVQRLDSRTLVAAGHGRSGFLLAPWTAEQIVSELVSVGAAS--------
Rv0415|M.tuberculosis_H37Rv PLVQRLDSRTLVAAGHGRSGFLLAPWTAEQIVSELVSVGAAS--------
MLBr_00299|M.leprae_Br4923 PLVHRLDNRTLVAAGHGRSGFLLAPWTAEQIMFELAAVGAQA--------
MMAR_0718|M.marinum_M PIVGRLDARTLVAAGLGRSGFLLAPWTAEQIVSELVPIGAHS--------
MUL_2805|M.ulcerans_Agy99 PIVGRLDARTLVAAGLGRSGFLLAPWTAEQIVSELVPIGAHS--------
MAV_4746|M.avium_104 PLVGRLDARTLVAAGHGRSGFLLAPWTAEQIVSELAPVGAHR--------
Mvan_0708|M.vanbaalenii_PYR-1 PLVGRLDERTLVAAGHGRSGFLLAPWTAESIAAELEGALVR---------
Mflv_0197|M.gilvum_PYR-GCK PLVGRLDERTLVAAGHGRSGFLLAPWTVEKIAAELEGAMVMEVAE-----
MSMEG_0791|M.smegmatis_MC2_155 PVVERVDERTVVAVGHGRNGFLLAPWTAERVAAELEVGVGAK--------
TH_2312|M.thermoresistible__bu PIVGRLDDRTLVAAGHGRSGFLLAPWTAEAILAELDGAPLPEAELVGAR-
MAB_4218c|M.abscessus_ATCC_199 PMVGRIDERVVVAAGHGRSGFLLAPWTAKTIGAVLDGGESPAAVDPRRFE
*:* *:* *.:**.* **.********.: : *
Mb0423|M.bovis_AF2122/97 --------
Rv0415|M.tuberculosis_H37Rv --------
MLBr_00299|M.leprae_Br4923 --------
MMAR_0718|M.marinum_M --------
MUL_2805|M.ulcerans_Agy99 --------
MAV_4746|M.avium_104 --------
Mvan_0708|M.vanbaalenii_PYR-1 --------
Mflv_0197|M.gilvum_PYR-GCK --------
MSMEG_0791|M.smegmatis_MC2_155 --------
TH_2312|M.thermoresistible__bu --------
MAB_4218c|M.abscessus_ATCC_199 KAHSQEAR