For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPIATPEVYAEMLGRAKEHSFAFPAINCVGSESVNAAIKGFADAGSDGIIQFSTGGAEFASGLGVKEMVT GAVALAEFAHVIAAKYDITVALHTDHCPKDKLDTYVRPLLAISAERVARGQNPLFQSHMWDGSAVPIDEN LSIAQELLKQAAAAKIILEIEIGVVGGEEDGVEAEINDKLYTTPEDFEKTIEALGAGEHGKYLLAATFGN VHGVYKPGNVVLKPEVLAEGQRVASAKLGLPAGSKPFDFVFHGGSGSLKSEIEDSLRYGVVKMNVDTDTQ YAFTRPIVGHMFTNYDGVLKVDGEVGNKKVYDPRSYFKKAEAGMSERVVEACKDLHSAGRSVSAG
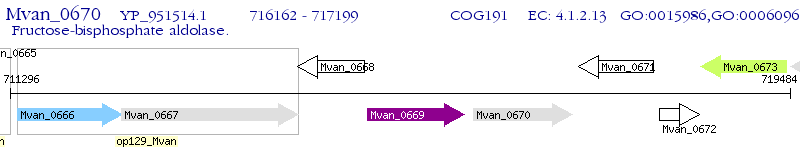
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0670 | - | - | 100% (345) | fructose-bisphosphate aldolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0370c | fba | 1e-175 | 86.59% (343) | fructose-bisphosphate aldolase |
| M. gilvum PYR-GCK | Mflv_0234 | - | 0.0 | 94.20% (345) | fructose-bisphosphate aldolase |
| M. tuberculosis H37Rv | Rv0363c | fba | 1e-175 | 86.59% (343) | fructose-bisphosphate aldolase |
| M. leprae Br4923 | MLBr_00286 | fba | 1e-170 | 84.01% (344) | fructose-bisphosphate aldolase |
| M. abscessus ATCC 19977 | MAB_4254c | - | 1e-175 | 87.83% (345) | fructose-bisphosphate aldolase |
| M. marinum M | MMAR_0669 | fba | 1e-175 | 86.01% (343) | fructose-bisphosphate aldolase, Fba |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0752 | fbaA | 0.0 | 92.44% (344) | fructose-bisphosphate aldolase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0103 | fba | 1e-175 | 85.71% (343) | fructose-bisphosphate aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0669|M.marinum_M -----MPIATPEVYAEMLGRAKENSYAFPAINCTSSETVNAALKGFADAG
MUL_0103|M.ulcerans_Agy99 -----MPIATPEVYAEMLGRAKENSYAFPAINCTSSETVNAALKGFADAG
Mb0370c|M.bovis_AF2122/97 -----MPIATPEVYAEMLGQAKQNSYAFPAINCTSSETVNAAIKGFADAG
Rv0363c|M.tuberculosis_H37Rv -----MPIATPEVYAEMLGQAKQNSYAFPAINCTSSETVNAAIKGFADAG
MLBr_00286|M.leprae_Br4923 -----MPIATPEIYAEMLRRAKENSYAFPAINCTSSETVNAAIKGFADAG
Mvan_0670|M.vanbaalenii_PYR-1 -----MPIATPEVYAEMLGRAKEHSFAFPAINCVGSESVNAAIKGFADAG
Mflv_0234|M.gilvum_PYR-GCK -----MPIATPEVYAEMLARAKEHSFAFPAINCVGSESVNAAIKGFADAG
MSMEG_0752|M.smegmatis_MC2_155 -----MPIATPEVYAEMLDRAKAHSFAFPAINCVGSESINAAIKGFADAG
MAB_4254c|M.abscessus_ATCC_199 MEVTQVPIATPEVYTEMLKRAKEHSFAFPAINCTSSETINAAIKGFADAG
:******:*:*** :** :*:*******..**::***:*******
MMAR_0669|M.marinum_M SDGIIQFSTGGAEFGSGLGIKNMVTGAVALAEFTHIVAAKYPINVALHTD
MUL_0103|M.ulcerans_Agy99 SDGIIQFSTGGAEFGSGLGIKNMVTGAVALAEFTHIVAAKYPINVALHTD
Mb0370c|M.bovis_AF2122/97 SDGIIQFSTGGAEFGSGLGVKDMVTGAVALAEFTHVIAAKYPVNVALHTD
Rv0363c|M.tuberculosis_H37Rv SDGIIQFSTGGAEFGSGLGVKDMVTGAVALAEFTHVIAAKYPVNVALHTD
MLBr_00286|M.leprae_Br4923 SDGIIQFSTGGAEFASGLGVKDMVTGAVALAKFTHTIAAKYPINVALHTD
Mvan_0670|M.vanbaalenii_PYR-1 SDGIIQFSTGGAEFASGLGVKEMVTGAVALAEFAHVIAAKYDITVALHTD
Mflv_0234|M.gilvum_PYR-GCK SDGIIQFSTGGAEFASGLGVKDMVTGAVALAEFAHVIAEKYPITVALHTD
MSMEG_0752|M.smegmatis_MC2_155 SDGIIQFSTGGAEFGSGLGVKDMVTGAVALAEFAHVVAAKYPITVALHTD
MAB_4254c|M.abscessus_ATCC_199 SDGIIQFSTGGAEFGSGLGVKDMVTGAVALAEFAHVVAAKYDITVALHTD
**************.****:*:*********:*:* :* ** :.******
MMAR_0669|M.marinum_M HCPKDKLDTYVRPLLAISAERVSAGKDPLFQSHMWDGSAVPIDENLVIAQ
MUL_0103|M.ulcerans_Agy99 HCPKDKLDTYVRPLLAISAERVSAGKDPLFQSHMWDGSAVPIDENLVIAQ
Mb0370c|M.bovis_AF2122/97 HCPKDKLDSYVRPLLAISAQRVSKGGNPLFQSHMWDGSAVPIDENLAIAQ
Rv0363c|M.tuberculosis_H37Rv HCPKDKLDSYVRPLLAISAQRVSKGGNPLFQSHMWDGSAVPIDENLAIAQ
MLBr_00286|M.leprae_Br4923 HCPKDKLDSYVRPLLAISARRVATGKDPLFGSHMWDGSAIPIDENLAIAQ
Mvan_0670|M.vanbaalenii_PYR-1 HCPKDKLDTYVRPLLAISAERVARGQNPLFQSHMWDGSAVPIDENLSIAQ
Mflv_0234|M.gilvum_PYR-GCK HCPKDKLDTYVRPLLAISSERVQAGRNPLFQSHMWDGSAVPIDENLSIAQ
MSMEG_0752|M.smegmatis_MC2_155 HCPKDKLDTYVRPLLAISAERVARGENPLFQSHMWDGSAVPIDENLTIAQ
MAB_4254c|M.abscessus_ATCC_199 HCPKDKLDTYVRPLLAISQERVNAGQAPLFQSHMWDGSAVPIDENLTIAA
********:********* .** * *** ********:****** **
MMAR_0669|M.marinum_M ELLKAAVAAKIILEIEIGVVGGEEDGVEAEINEKLYTTPEDFEKTVDALG
MUL_0103|M.ulcerans_Agy99 ELLKAAVAAKIILEIEIGVVGGEEDGVEAEINEKLYTTPEDFEKTVDALG
Mb0370c|M.bovis_AF2122/97 ELLKAAAAAKIILEIEIGVVGGEEDGVANEINEKLYTSPEDFEKTIEALG
Rv0363c|M.tuberculosis_H37Rv ELLKAAAAAKIILEIEIGVVGGEEDGVANEINEKLYTSPEDFEKTIEALG
MLBr_00286|M.leprae_Br4923 DLLKDAAAAKIILEVEIGVVGGEEDGVAGEINEKLYTTPKDFVKTIDALG
Mvan_0670|M.vanbaalenii_PYR-1 ELLKQAAAAKIILEIEIGVVGGEEDGVEAEINDKLYTTPEDFEKTIEALG
Mflv_0234|M.gilvum_PYR-GCK ELLKQAAAAKIILEIEIGVVGGEEDGVEAEINDKLYTTPEDFEKTIEALG
MSMEG_0752|M.smegmatis_MC2_155 ELLKTAAAAKIILEVEIGVVGGEEDGVEAEINDKLYTSPEDFEKTIAALG
MAB_4254c|M.abscessus_ATCC_199 ELLSAAVKAKIILEVEIGVVGGEEDGVEAEINEKLYTSSEDFEKTVDALG
:**. *. ******:************ ***:****:.:** **: ***
MMAR_0669|M.marinum_M AGEQGTYLLAATFGNVHGVYKPGNVKLRPDILDQGQKVAAAKLGLPDSAK
MUL_0103|M.ulcerans_Agy99 AGEQGTYLLAATFGNVHGAYKPGNVKLRPDILDQGQKVAAAKLGLPASAK
Mb0370c|M.bovis_AF2122/97 AGEHGKYLLAATFGNVHGVYKPGNVKLRPDILAQGQQVAAAKLGLPADAK
Rv0363c|M.tuberculosis_H37Rv AGEHGKYLLAATFGNVHGVYKPGNVKLRPDILAQGQQVAAAKLGLPADAK
MLBr_00286|M.leprae_Br4923 AGEHGKYLLAATFGNVHGVYKPGNVKLRPDILAEGQKVAAAKLSQSEGSK
Mvan_0670|M.vanbaalenii_PYR-1 AGEHGKYLLAATFGNVHGVYKPGNVVLKPEVLAEGQRVASAKLGLPAGSK
Mflv_0234|M.gilvum_PYR-GCK VGEHGKYLLAATFGNVHGVYKPGNVKLKPEVLAEGQKVASAKLGLEDGSK
MSMEG_0752|M.smegmatis_MC2_155 AGEEGRYLLAATFGNVHGVYKPGNVVLKPEVLAEGQRVAAAKLGLPADAK
MAB_4254c|M.abscessus_ATCC_199 AGDKGHYLLAATFGNVHGVYKPGNVKLKPEVLAEGQKVASAKLGLPEGSK
.*:.* ************.****** *:*::* :**:**:***. .:*
MMAR_0669|M.marinum_M PFDFVFHGGSGSLKSEIEEALRYGVVKMNVDTDTQYAFSRPIVGHMFTNY
MUL_0103|M.ulcerans_Agy99 PFDFVFHGGSGSLKSEIEEALRYGVVKMNVDTDTQYAFSRPIVGHIFTNY
Mb0370c|M.bovis_AF2122/97 PFDFVFHGGSGSLKSEIEEALRYGVVKMNVDTDTQYAFTRPIAGHMFTNY
Rv0363c|M.tuberculosis_H37Rv PFDFVFHGGSGSLKSEIEEALRYGVVKMNVDTDTQYAFTRPIAGHMFTNY
MLBr_00286|M.leprae_Br4923 PFDFVFHGGSGSEKSEIEEALRYGVVKMNVDTDTQYAFTRPVSGHMFTNY
Mvan_0670|M.vanbaalenii_PYR-1 PFDFVFHGGSGSLKSEIEDSLRYGVVKMNVDTDTQYAFTRPIVGHMFTNY
Mflv_0234|M.gilvum_PYR-GCK PFDFVFHGGSGSLKSEIEDSLRYGVVKMNVDTDTQYAFTRPVAAHMFSNY
MSMEG_0752|M.smegmatis_MC2_155 PFDFVFHGGSGSLKSEIEDSLKYGVVKMNVDTDTQYAFTRPIAGHMFTNY
MAB_4254c|M.abscessus_ATCC_199 PFDFVFHGGSGSLKSEIEDSLRYGVVKMNVDTDTQYAFTRPIAAHMFSNY
************ *****::*:****************:**: .*:*:**
MMAR_0669|M.marinum_M DGVLKVDGEVGNKKVYDPRSYLKKAEAGMTERVIEACNDLHCAGKSLAG-
MUL_0103|M.ulcerans_Agy99 DGVLKVDGEVGNKKVYDPRSYLKKAEAGMTERVIEACNDLHCAGKSLAG-
Mb0370c|M.bovis_AF2122/97 DGVLKVDGEVGVKKVYDPRSYLKKAEASMSQRVVQACNDLHCAGKSLTH-
Rv0363c|M.tuberculosis_H37Rv DGVLKVDGEVGVKKVYDPRSYLKKAEASMSQRVVQACNDLHCAGKSLTH-
MLBr_00286|M.leprae_Br4923 DGVLKVDGDVGNKKVYDPRSYLKKAEASMTERVLEACNDLRCAGKSVAAS
Mvan_0670|M.vanbaalenii_PYR-1 DGVLKVDGEVGNKKVYDPRSYFKKAEAGMSERVVEACKDLHSAGRSVSAG
Mflv_0234|M.gilvum_PYR-GCK DGVLKVDGEVGNKKVYDPRSYFKKAEASMAERVVEACNDLHSAGRSVSAG
MSMEG_0752|M.smegmatis_MC2_155 DGVLKIDGEVGNKKTYDPRSYLKKAEASMSERVVEACNDLHSAGRSVTAA
MAB_4254c|M.abscessus_ATCC_199 DGVLKIDGEVGNKKTYDPRSYLKKAEANMSVRVVEACNDLKSAGRSVSAG
*****:**:** **.******:*****.*: **::**:**:.**:*::
MMAR_0669|M.marinum_M --
MUL_0103|M.ulcerans_Agy99 --
Mb0370c|M.bovis_AF2122/97 --
Rv0363c|M.tuberculosis_H37Rv --
MLBr_00286|M.leprae_Br4923 --
Mvan_0670|M.vanbaalenii_PYR-1 --
Mflv_0234|M.gilvum_PYR-GCK --
MSMEG_0752|M.smegmatis_MC2_155 --
MAB_4254c|M.abscessus_ATCC_199 LA