For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHGPHDLGGRDGIGPVDPEFLKPGIPEWRYPFEGRMHAVTAMAIAKGAFNLDEQRHGIELMKWSDYLDST YYAHWLFSVEKLLNDKGYITHEEIDERMRELGTPSMTPHPTRPETLSPFAEKMIEVLWAGTPHDRPTDQP AAYAVGDAVMVRNLNEATHNRLPGYLNKTPGVIAKHYGAHHDPADSAHRQVETPHHLYMVRFTSTDLWGD EAPEAFDLYADLFENYLEPA
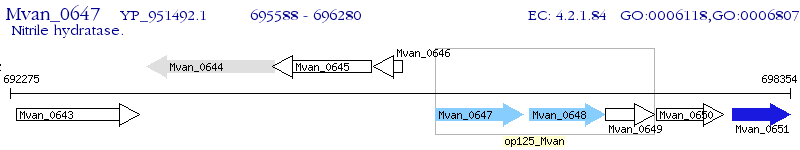
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0647 | - | - | 100% (230) | nitrile hydratase |
| M. vanbaalenii PYR-1 | Mvan_5826 | - | 3e-17 | 31.25% (176) | nitrile hydratase |
| M. vanbaalenii PYR-1 | Mvan_1505 | - | 4e-08 | 28.30% (159) | hypothetical protein Mvan_1505 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1010 | - | 8e-20 | 32.95% (176) | nitrile hydratase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0377 | - | 1e-19 | 28.69% (244) | nitrile hydratase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1010|M.gilvum_PYR-GCK MKLQHYLGGLEGLPEPLNLEKKVFVEDWEKRIFGIHVAMMGLSNHLGSAL
MSMEG_0377|M.smegmatis_MC2_155 MRLQHFIGGLEGLTGPLDFEKRVFVEDWEKRIFGIHVAMMALSTHLGEAL
Mvan_0647|M.vanbaalenii_PYR-1 MHGPHDLGGRDGIGPVDPEFLKPGIPEWRYPFEGRMHAVTAMAIAKG---
*: * :** :*: : : :*. : * *: .:: *
Mflv_1010|M.gilvum_PYR-GCK PGYPIAEVPTTFRDEWTWASLRTGAEGMNPFDYFKFRYYEKWLGGISQFF
MSMEG_0377|M.smegmatis_MC2_155 PGYRVADVPTTFREKWTWAHLRTGAEGMNPFDYFKFRYYEKWLGGISQFF
Mvan_0647|M.vanbaalenii_PYR-1 --------------AFNLDEQRHGIELMKWSDYLDSTYYAHWLFSVEKLL
:. * * * *: **:. ** :** .:.:::
Mflv_1010|M.gilvum_PYR-GCK VDNGYVSEEEIADLLGVT-----DPPVAGPGS-PAIDEQVIDYLRRGDSP
MSMEG_0377|M.smegmatis_MC2_155 VDKGYLSEDEIRAAPVGS-----PPKAPAPGVKAAIDDQVLDYLRRGDSP
Mvan_0647|M.vanbaalenii_PYR-1 NDKGYITHEEIDERMRELGTPSMTPHPTRPETLSPFAEKMIEVLWAGTPH
*:**::.:** * . * ..: ::::: * * .
Mflv_1010|M.gilvum_PYR-GCK RRDVAHP-KFEVGDTVLITNAPAGAHTRLPGYLRAKTGTVQRIFEGDYGY
MSMEG_0377|M.smegmatis_MC2_155 RRGSAIP-RFAPGDKVRIADVPAGEHTRLPGYLRNRTGTVERCFEGEYSY
Mvan_0647|M.vanbaalenii_PYR-1 DRPTDQPAAYAVGDAVMVRNLNEATHNRLPGYLNKTPGVIAKHYGAHHDP
* * : ** * : : . *.******. .*.: : : ..:.
Mflv_1010|M.gilvum_PYR-GCK FVHTGDGIGDPMPIYIVEFTPEELWGPRAEPGANALYAELFEAYLEPIEE
MSMEG_0377|M.smegmatis_MC2_155 FVHTGDGIGDPMPIYIVEFTPRELWGARAEPGASALYAELFEAYLSPVEE
Mvan_0647|M.vanbaalenii_PYR-1 ADSAHRQVETPHHLYMVRFTSTDLWG-DEAPEAFDLYADLFENYLEPA--
: : * :*:*.**. :*** * * ***:*** **.*
Mflv_1010|M.gilvum_PYR-GCK DK
MSMEG_0377|M.smegmatis_MC2_155 DQ
Mvan_0647|M.vanbaalenii_PYR-1 --