For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
THTHSHSHSLNGPSPLGPLAARIVVGLLVAIGVATLIGAALLWPSGGKADIPLPFQNAAGGAVTTESGHV LSSSAAVCGSPSVGAVLTSAPVPAEGEGAACVQALVAIDSGPNQGANTLLEFSGGPGQPSLAVGDDIRVS RQVDDVGSTTYAFYDFERAWPLTVLAAVFAIVIVAVARWRGLRALVGIVIAFGVLVVFLLPALRDGAPAV PVSLVASAAILYAVIYLAHGVSLRTSAALLGTLASLLLAAVLSWAAIELAHLTGLSEDQNNTVAAYLGNV SITGLLLAGFIIGSLGVLNDVTVTQASTAFELAALGDGASRRAVFVGAMRVGRDHIASTVYTLVLAYAGS ALPLLLLFSVANRSLGDVLTSESVAIEIARSAVGGIALALSVPLTTGIAVLLATPRHTSH*
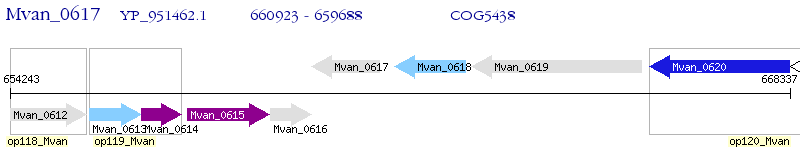
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0617 | - | - | 100% (411) | YibE/F family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0281 | - | 0.0 | 87.35% (411) | YibE/F family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1599 | - | 1e-154 | 72.82% (379) | hypothetical protein MAB_1599 |
| M. marinum M | MMAR_0379 | pntB | 8e-06 | 24.21% (285) | NAD(P) transhydrogenase (subunit beta) PntB |
| M. avium 104 | MAV_4819 | - | 1e-167 | 76.98% (404) | hypothetical protein MAV_4819 |
| M. smegmatis MC2 155 | MSMEG_0687 | - | 0.0 | 83.25% (406) | hypothetical protein MSMEG_0687 |
| M. thermoresistible (build 8) | TH_0765 | - | 1e-180 | 81.48% (405) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0617|M.vanbaalenii_PYR-1 ------MTHTHSHSHSLNGPSPLGPLAARIVVGLLVAIGVATLIGAALLW
Mflv_0281|M.gilvum_PYR-GCK ----MRVAHSHSHTHSLSGPSPLGPLAAKVVVGLLVVIGVAVMIGAALLW
TH_0765|M.thermoresistible__bu ------VTHSHSHSH--TGPAPLGPPAARLVVAALVLIALATVAGAVLLW
MSMEG_0687|M.smegmatis_MC2_155 ------MAHSHSHSHSLSGPSPLGPLAARIVVGLLVAIGVVVIVGAALLW
MAV_4819|M.avium_104 ------MTHSHSHGLP-SGPAPIDRLPARIVVGLLIAIGVAVAVGAIVLW
MAB_1599|M.abscessus_ATCC_1997 ---------------------------------MLAAMGIAVLVGLVALW
MMAR_0379|M.marinum_M MNYLVIGLYILAFSLFIYGLMGLTGPKTAVRGNLIAAVGMAIAVAATLVK
: :.:. . :
Mvan_0617|M.vanbaalenii_PYR-1 PSGGKADIPLPFQNAAGGAVTTESGHVLSSSAAVCGSPSVGAVLTSAPVP
Mflv_0281|M.gilvum_PYR-GCK PDGQKSDIPLPFQNAAGGAVTTESGHVLSSVAATCGSPSAGSVLTSAPLP
TH_0765|M.thermoresistible__bu PSGEKAEIPLPFQDATGGAVTTEAGRVLSSTLATCGSPSVGAVLTTTPLP
MSMEG_0687|M.smegmatis_MC2_155 PSRQKVDIPLPYQNAAGGAVTTETGHVLSSTLTACGSPSVGAVLTSDPVS
MAV_4819|M.avium_104 PSRQHVDIPMPLQNAAGGAVSTQAGHVLSSGLGDCGSPSVSQVLTGPPQP
MAB_1599|M.abscessus_ATCC_1997 PEHRTVDIPLPYQNAHGGAVTTESGTVLATRLGECGSPSTGQVLTADPAA
MMAR_0379|M.marinum_M IRHTDQWVLIIAGLVVGVALGVPPARLTKMTAMPQLVAFFNGVGGGTVAL
: : . * *: . .. : . . *
Mvan_0617|M.vanbaalenii_PYR-1 AEGEGAACVQALVAIDSGPNQGANTLLEFSGGPGQPSLAVGDDIRVSRQV
Mflv_0281|M.gilvum_PYR-GCK AEGGGAECVQSLVSIDSGPNEGADTLLEIGVGPGQPSLAVGDDVRLSRQV
TH_0765|M.thermoresistible__bu APGDGGQCVQNLIAIESGPNSGARTVLEFSGGPGQPELAPGDEIRISRQV
MSMEG_0687|M.smegmatis_MC2_155 SPGDAGQCVQNLVAIDSGPNKGARTLLEFSGGPGQPNLVAGDDVRLSRQV
MAV_4819|M.avium_104 ALPGAGRCVLTQVAIDSGPNAGAATLLESSPGPGQPKFAVGDRIRIVRQV
MAB_1599|M.abscessus_ATCC_1997 GQTPDSACALDLIAITSGPNSGAKTQLEFTAGGGQPTLAAGEHIRITRQV
MMAR_0379|M.marinum_M IALSEFIETKGFSAFQHGESPTVHIVVASLFAAIIGSISFWGSIIAFGKL
. :: * . . : . : : ::
Mvan_0617|M.vanbaalenii_PYR-1 D-DVGSTTYAFYDFERAWPLTVLA-AVFAIVIVAVARWRG-----LRALV
Mflv_0281|M.gilvum_PYR-GCK D-DAGTTTYAFYDFERTWPLTAFA-LVFAIVIVAVARWRG-----LRALV
TH_0765|M.thermoresistible__bu DGDTGVTTYAFYDYARGLPLAVLA-AAFAVVIVAVARWRG-----LRALL
MSMEG_0687|M.smegmatis_MC2_155 D-ASGATSYAFYDYERTWPLIALA-AVFAVVVVAVARWRG-----LRALI
MAV_4819|M.avium_104 D-DQGATSYAFYDFERGWALVGLA-VAFAVVIVAVARWRG-----LLALV
MAB_1599|M.abscessus_ATCC_1997 D-AQGATSYSFYDYERTWPLIAVA-AAFAIVICLVARWRG-----LRALV
MMAR_0379|M.marinum_M QELISGAPIGFGKAQQPVNLLLLVGAVVAAVVIGLDAHPGSGGVSLWWMI
: . :. .* . : * .. ..* *: : * * ::
Mvan_0617|M.vanbaalenii_PYR-1 GIVIAFGVLVVFLLPALRDGAPAVPVSLV---------ASAAILYAVIYL
Mflv_0281|M.gilvum_PYR-GCK GIVIAFGVLVVFLLPALRDGAPAVPVALV---------ASAAILYAVLYL
TH_0765|M.thermoresistible__bu GIVIAFGVLVVFLLPALRDGAPAVPVALV---------GSAAILFAVLYL
MSMEG_0687|M.smegmatis_MC2_155 GIVVAFGVLVVFLLPALRDGASAVPVSLV---------ASAVILYAVIYL
MAV_4819|M.avium_104 GILVAFVVLVTFLLPALRDGAPAVPVALV---------AAAAILYAVIYL
MAB_1599|M.abscessus_ATCC_1997 GILVAFLVLAVFLLPALRDGGPALPLALI---------ASAAILFVVIYL
MMAR_0379|M.marinum_M GLLAAAGVLGLMVVLPIGGADMPVVISLLNAMTGLSAAAAGLALNNTAMI
*:: * ** ::: .: .. .: ::*: .:. * . :
Mvan_0617|M.vanbaalenii_PYR-1 AHGVSLRTSAALLGTLASLLLAAVLSWAAIELAHLTGLSEDQNNTVAAYL
Mflv_0281|M.gilvum_PYR-GCK AHGVSLRTSAALLGTLASLLLAAGLSWAAIEFTHITGLSEDQNNEVAAYL
TH_0765|M.thermoresistible__bu AHGVSLRTSAALLGTLMSLLLAAFLSWAAIELLHLTGLSEDQNNEVAAYL
MSMEG_0687|M.smegmatis_MC2_155 AHGVSLRTSAALLGTLTALFLAALLSWAAIELAHLTGLAEDQNNDVAAYM
MAV_4819|M.avium_104 AHGVSLRTSAALLGTLSSLLLAAGLSWAAIQLAHLTGLSDEQNSTVSAYL
MAB_1599|M.abscessus_ATCC_1997 AHGVSMRTSAALLGTLTAMLLAAGLSWAVIELTHLTGLSDEQNNEVAAYM
MMAR_0379|M.marinum_M VAGMIVGASGSILTNLMAKAMNRSIPAIVAGGFGGGGVSPSADGGGDKHV
. *: : :*.::* .* : : :. . *:: . :. ::
Mvan_0617|M.vanbaalenii_PYR-1 GNVSITGLLLAGFIIGSLGVLNDVTVTQASTAFELAALG-----DGASRR
Mflv_0281|M.gilvum_PYR-GCK GNVSITGMLLAGFIIGSLGVLNDVTVTQASTAFELAAVG-----DGATRR
TH_0765|M.thermoresistible__bu GNVSITGLLLAGFIIGSLGVLNDVTITQASAVFELANL------EGSSRR
MSMEG_0687|M.smegmatis_MC2_155 GNVSITGLLLAGFIIGSLGVLNDVTVTQASAVFELAEA-------GGTRR
MAV_4819|M.avium_104 GSVSIGGLLLAGFIIGSLGVLNDVTVTQSSTVFELARL-------GGSRR
MAB_1599|M.abscessus_ATCC_1997 GGVSISGLLLAGFIIGSLGVLNDVTVTQASTVYELAELG-----DGSSRR
MMAR_0379|M.marinum_M KATSAADAAIQMAYANQVIVVPGYGLAVAQAQHAVKDMASLLEDKGVAVK
.* . : ..: *: . :: :.: . : * : :
Mvan_0617|M.vanbaalenii_PYR-1 AVFVGAMRVGRDHIASTVYTLVLAYAGSALPLLLLFSVANRSLGDVLTSE
Mflv_0281|M.gilvum_PYR-GCK QVFVGAMRVGRDHIASTVYTLVLAYAGSALPLLLLFSVANRSLGDVLTSE
TH_0765|M.thermoresistible__bu AAFVGAMRVGSDHIASTVYTLVLAYAGSALPLLLLFSVADRSLTDVLTSE
MSMEG_0687|M.smegmatis_MC2_155 SIFVGAMRVGRDHIASTVYTLVLAYAGSALPLLLLFSVANRSLGDVLTSE
MAV_4819|M.avium_104 AIFTGALRVGRDHIASTVYTLVLAYAGSSLPLLLLFSVANRSLTDVLTGE
MAB_1599|M.abscessus_ATCC_1997 DIFLGAMRVGRDHIASTVYTLVLAYAGNSLPLMLLFSVANRSLGDVLTSE
MMAR_0379|M.marinum_M YAIHPVAGRMPGHMNVLLAEAEVDYDAMKDMDDINDEFARTDVAIVIGAN
: . .*: : : * . : ..* .: *: .:
Mvan_0617|M.vanbaalenii_PYR-1 SVAIEIARSAVGGIALALSVPLTTGIAVLLATPRHTS---H---------
Mflv_0281|M.gilvum_PYR-GCK SVAIEVARSAVGGIALALSVPLTTGIAAMLATPQQRR---H---------
TH_0765|M.thermoresistible__bu LVAIEVARSAVGGIALALSVPLTTAIAAVLATPKSRAGAPHSTRSASVSP
MSMEG_0687|M.smegmatis_MC2_155 NVAVEVARSAVGGIALALSVPLTTGIAAVLATPSVASPAR----------
MAV_4819|M.avium_104 SVAIEIARSAVGGMALALSVPLTTAIAAVLARPSGGRKRGEAGRGGPPPS
MAB_1599|M.abscessus_ATCC_1997 GVAIEIARSAVGGIALVLSVPLTTAIAAALATPGKVRTGA----------
MMAR_0379|M.marinum_M DVTNPAARNDASSPIYGMPILNVDKAKSVIVLKRSMNSGFAGIDNPLFYG
*: **. ... :.: . :.
Mvan_0617|M.vanbaalenii_PYR-1 ------------------------
Mflv_0281|M.gilvum_PYR-GCK ------------------------
TH_0765|M.thermoresistible__bu VARRAEPRSS--------------
MSMEG_0687|M.smegmatis_MC2_155 ------------------------
MAV_4819|M.avium_104 V-----------------------
MAB_1599|M.abscessus_ATCC_1997 ------------------------
MMAR_0379|M.marinum_M EGTTMLFGDAKKSVTEVAEELKAL