For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LPSRNLDKNVSMPVEQNTKVAIVGMGSVGTAIAYACLIRGSAGALALYDVNSTKVRAEVLDLKHGSQFVP HCRISGSDDIAVTAGSAVVIVTAGAKQKPGQSRLELAAANVAMAQTLTPQLLAHSPEAVVIFVTNPVDVV TFAATRSVDAAPGHVFGSGTVLDSSRFRYLIAQQADLAVGNVHGLIVGEHGDSEISLWSSVSVGGVPATE FRRDGVLVFDEQNRRRISTDVVNAAYEIIAGKGATNLAIGLSSARIIEAVLGDQHRVLPVSTVQQGAHGI SGVALSLPTVVSAHGAGQVLEVPLSESESQGLQASATTLRAAQESLGL
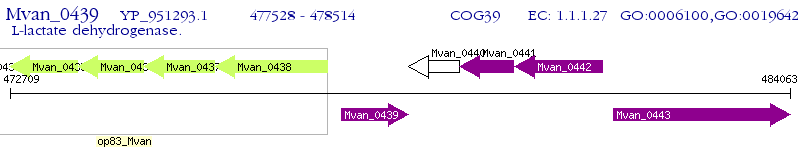
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0439 | - | - | 100% (328) | L-lactate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3067 | - | 2e-05 | 23.64% (330) | malate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0307 | - | 1e-153 | 85.80% (317) | L-lactate dehydrogenase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0439|M.vanbaalenii_PYR-1 -----------MPSRNLDKNVS-----MPVEQNTKVAIVGMGSVGTAIAY
Mflv_0307|M.gilvum_PYR-GCK MFSAVGSIGCGGDSRSTDILLWGEDGPMPVEQNTKVSVIGMGSVGTAIAY
**. * : *********:::***********
Mvan_0439|M.vanbaalenii_PYR-1 ACLIRGSAGALALYDVNSTKVRAEVLDLKHGSQFVPHCRISGSDDIAVTA
Mflv_0307|M.gilvum_PYR-GCK ACLIRGSAGALALYDLNAAKVRAEVLDLNHGSQFVPHCRITGSDDIAVTD
***************:*::*********:***********:********
Mvan_0439|M.vanbaalenii_PYR-1 GSAVVIVTAGAKQKPGQSRLELAAANVAMAQTLTPQLLAHSPEAVVIFVT
Mflv_0307|M.gilvum_PYR-GCK GSAVIVVTAGAKQKPGQSRLELAATNVAMAQTLTPQLLERSPDAVIVFVT
****::******************:************* :**:**::***
Mvan_0439|M.vanbaalenii_PYR-1 NPVDVVTFAATRSVDAAPGHVFGSGTVLDSSRFRYLIAQQADLAVGNVHG
Mflv_0307|M.gilvum_PYR-GCK NPVDVVTFAAARSVNAAPGHIFGSGTVLDSSRFRYLIADRADLAVANVHG
**********:***:*****:*****************::*****.****
Mvan_0439|M.vanbaalenii_PYR-1 LIVGEHGDSEISLWSSVSVGGVPATEFRRDGVLVFDEQNRRRISTDVVNA
Mflv_0307|M.gilvum_PYR-GCK IIVGEHGDSEISLWSSVSVGGVPAAQFRRDGVPVFDEDTRKRISAEVVNA
:***********************::****** ****:.*:***::****
Mvan_0439|M.vanbaalenii_PYR-1 AYEIIAGKGATNLAIGLSSARIIEAVLGDQHRVLPVSTVQQGAHGISGVA
Mflv_0307|M.gilvum_PYR-GCK AYEIIAGKGATNLAIGLSTARIVEAVLGDEHRVLPVSTVQEGAYGILGVA
******************:***:******:**********:**:** ***
Mvan_0439|M.vanbaalenii_PYR-1 LSLPTVVSAHGAGQVLEVPLSESESQGLQASATTLRAAQESLGL
Mflv_0307|M.gilvum_PYR-GCK LSLPTVVSAHGAGRVLEVALSEDERVGLQASATTLRQAQERLGL
*************:****.***.* ********** *** ***