For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FSAVGSIGCGGDSRSTDILLWGEDGPMPVEQNTKVSVIGMGSVGTAIAYACLIRGSAGALALYDLNAAKV RAEVLDLNHGSQFVPHCRITGSDDIAVTDGSAVIVVTAGAKQKPGQSRLELAATNVAMAQTLTPQLLERS PDAVIVFVTNPVDVVTFAAARSVNAAPGHIFGSGTVLDSSRFRYLIADRADLAVANVHGIIVGEHGDSEI SLWSSVSVGGVPAAQFRRDGVPVFDEDTRKRISAEVVNAAYEIIAGKGATNLAIGLSTARIVEAVLGDEH RVLPVSTVQEGAYGILGVALSLPTVVSAHGAGRVLEVALSEDERVGLQASATTLRQAQERLGL*
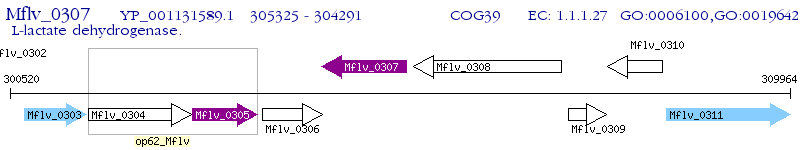
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0307 | - | - | 100% (344) | L-lactate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01091 | mdh | 6e-05 | 23.28% (262) | malate dehydrogenase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4198 | mdh | 2e-05 | 23.05% (269) | malate dehydrogenase Mdh |
| M. avium 104 | MAV_1380 | - | 3e-05 | 21.93% (269) | malate dehydrogenase |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4504 | mdh | 2e-06 | 23.55% (276) | malate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0439 | - | 1e-153 | 85.80% (317) | L-lactate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4198|M.marinum_M -----------MSAS-----------PLKVAVTGAAGQIGYSLLFRLASG
MUL_4504|M.ulcerans_Agy99 -----------MSAS-----------PLKVAVTGAAGQIGYSLLFRLASG
MAV_1380|M.avium_104 -----------MSAS-----------PLKVAVTGAAGQIGYSLLFRLASG
MLBr_01091|M.leprae_Br4923 -----------MSPR-----------PLKVAVTGAAGQIGYSLLFRLASG
Mflv_0307|M.gilvum_PYR-GCK MFSAVGSIGCGGDSRSTDILLWGEDGPMPVEQNTKVSVIGMGSVGTAIAY
Mvan_0439|M.vanbaalenii_PYR-1 -----------MPSRNLDKNVS-----MPVEQNTKVAIVGMGSVGTAIAY
. : * . .. :* . : :
MMAR_4198|M.marinum_M SLLGPDRPIELRLLEIEPALKALEGVVMELDDCAFPLLSGVEIGSDANKI
MUL_4504|M.ulcerans_Agy99 SLLGPDRPIELRLLEIEPALKALEGVVMELDDCAFPLLSGVEIGSDANKI
MAV_1380|M.avium_104 SLLGPDRPIELRLLEIEPALKALEGVVMELDDCAFPLLSGVEIGADPNKI
MLBr_01091|M.leprae_Br4923 SLLGLDRPIELRLLEIEPALKALEGVVMELDDCAFLLLAGVEIGADPNKV
Mflv_0307|M.gilvum_PYR-GCK ACLIRGSAGALALYDLN--AAKVRAEVLDLNHGSQFVPHCRITGSDDIAV
Mvan_0439|M.vanbaalenii_PYR-1 ACLIRGSAGALALYDVN--STKVRAEVLDLKHGSQFVPHCRISGSDDIAV
: * . . * * ::: :.. *::*.. : : *:* :
MMAR_4198|M.marinum_M FDGANLALLVGARPRGPGMERSDLLEANGAIFTAQGKALNEVAADDIRVG
MUL_4504|M.ulcerans_Agy99 FDGANLALLVGARPRGPGMERSDLLEANGAIFTAQGKALNEVAADDIRVG
MAV_1380|M.avium_104 FDGANLALLVGARPRGPGMERSDLLEANGAIFTAQGKALNEVAADDIRVG
MLBr_01091|M.leprae_Br4923 FDGVNLALLVGARPRGPGMERGDLLEANGAIFTAQGKALNAVAAADIRVG
Mflv_0307|M.gilvum_PYR-GCK TDGSAVIVVTAGAKQKPGQSRLELAATNVAMAQTLTPQLLERSPDAVIVF
Mvan_0439|M.vanbaalenii_PYR-1 TAGSAVVIVTAGAKQKPGQSRLELAAANVAMAQTLTPQLLAHSPEAVVIF
* : ::... : ** .* :* :* *: : * :. : :
MMAR_4198|M.marinum_M VTGNPANTNALIAMTNAPDIPRERFSALTRLDHNRAISQLAAKTGVAVTD
MUL_4504|M.ulcerans_Agy99 VTGNPANTNALIAMTNAPDIPRERFSALTRLDHNRAISQLAAKTGVAVTD
MAV_1380|M.avium_104 VTGNPANTNALIAMSNAPDIPRERFSALTRLDHNRAISQLAKKTGAKVTD
MLBr_01091|M.leprae_Br4923 VTGNPANTNALIAMTNAPDIPRERFSALTRLDHNRAIAQLATKTGSAVTD
Mflv_0307|M.gilvum_PYR-GCK VT-NPVDVVTFAAARSVNAAPGHIFGSGTVLDSSRFRYLIADRADLAVAN
Mvan_0439|M.vanbaalenii_PYR-1 VT-NPVDVVTFAATRSVDAAPGHVFGSGTVLDSSRFRYLIAQQADLAVGN
** **.:. :: * .. * . *.: * ** .* :* ::. * :
MMAR_4198|M.marinum_M IKKMTIWGNHSATQYPDLFHAEVKGKNAAEVVND-----QAWIEEYFIPT
MUL_4504|M.ulcerans_Agy99 IKKMTIWGNHSATQYPDLFHAEVKGKNAAEVVND-----QAWIEEYFIPT
MAV_1380|M.avium_104 ITKMTIWGNHSATQYPDIFHAEVKGKNAAEVVGD-----QNWIENDFIPT
MLBr_01091|M.leprae_Br4923 IRKMTIWGNHSATQYPDVFHAEIGGKNAAEVVGD-----QAWIEDYFIPT
Mflv_0307|M.gilvum_PYR-GCK VHGIIVG-EHGDSEISLWSSVSVGGVPAAQFRRDGVPVFDEDTRKRISAE
Mvan_0439|M.vanbaalenii_PYR-1 VHGLIVG-EHGDSEISLWSSVSVGGVPATEFRRDGVLVFDEQNRRRISTD
: : : :*. :: . ..: * *::. * : . : .
MMAR_4198|M.marinum_M VAKRGAAIIDARGASSAASAASATVDAARSWLLGTPADDWVSMAVLSDGS
MUL_4504|M.ulcerans_Agy99 VAKRGATIIDARGASSAASAASASVDAARSWLLGTPADDWVSMAVLSDGS
MAV_1380|M.avium_104 VAKRGAAIIDARGASSAASAASATTDAARDWLLGTPAGDWVSMAVISDGS
MLBr_01091|M.leprae_Br4923 VAKRGAAIIDARGASSAASAASATIDAARDWLLGTPEGNWVSMAVVSDGS
Mflv_0307|M.gilvum_PYR-GCK VVNAAYEIIAGKGATNLAIGLSTAR--IVEAVLGDEHR-VLPVSTVQEGA
Mvan_0439|M.vanbaalenii_PYR-1 VVNAAYEIIAGKGATNLAIGLSSAR--IIEAVLGDQHR-VLPVSTVQQGA
*.: . ** .:**:. * . *:: . :** :.::.:.:*:
MMAR_4198|M.marinum_M YGVPEGLISSFPVTTKDGNWSIVKGLEIDEFSRGRIDKTTAELADERKAV
MUL_4504|M.ulcerans_Agy99 YGVPEGLISSFPVTTKDGNWSIVKGLEIDEFSRGRIDKTAAELADERKAV
MAV_1380|M.avium_104 YGVPEGLISSFPVTTKDGDWTIVQGLEIDEFSRSRIDKTTAELADERNAV
MLBr_01091|M.leprae_Br4923 YGVPEGLISSFPVTTTDGDWTIVRGLGIDDFSRGRIDKSTAELADERMAV
Mflv_0307|M.gilvum_PYR-GCK YGILG-VALSLPTVVSAHGAGRVLEVALSEDERVGLQASATTLR---QAQ
Mvan_0439|M.vanbaalenii_PYR-1 HGISG-VALSLPTVVSAHGAGQVLEVPLSESESQGLQASATTLR---AAQ
:*: : *:*.... . * : :.: . :: ::: * *
MMAR_4198|M.marinum_M TELGLI
MUL_4504|M.ulcerans_Agy99 TELGLI
MAV_1380|M.avium_104 TQLGLI
MLBr_01091|M.leprae_Br4923 KQLGLI
Mflv_0307|M.gilvum_PYR-GCK ERLGL-
Mvan_0439|M.vanbaalenii_PYR-1 ESLGL-
***